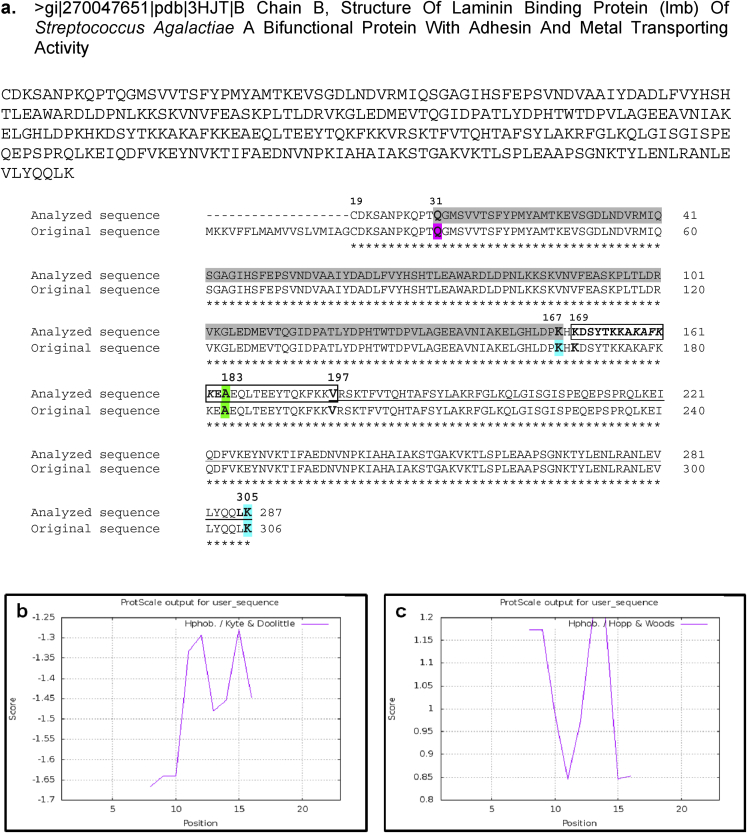
Fig. 2.
Structure of the Lmb mimicked MOSPD2 antigen.(a) The FASTA sequence is the analyzed Lmb sequence used in this study with 287 residues (20–306 amino acids). The original sequence of Lmb protein was obtained from the crystallography study with pdb id 3HJT (Ragunathan, 2009) containing 306 residues. The blocked region from 169-197 is the rigid linker helix containing the desired Lmb peptide sequence 169-‘KDSYTKKAKAFKKEA’-183 in bold in which the human motile sperm domain-containing protein 2 (MOSPD2) mimicked portion ‘KAFKK’ is represented in italics. The Lmb mimicked MOSPD2 peptide span over 169–183 amino acid length and is located in the α-helix of the entire Lmb protein of S. agalactiae. The numbers in bold represented the start and end positions of each region. Hydropathy plots for the Lmb mimicked MOSPD2 antigen using ExPasy Protscale. Hydropathy plots using ExPASy provided ProtScale tool are shown here. Basically, two different tools namely (b) Kyte Doolittle and (c) Hope-Woods hydropathy plots were used to cross-check the peptide properties before synthesis. The Kyte-Doolittle and the Hopp-Woods scale measures hydrophobic and hydrophilic residues above the scale bar 0, respectively (Kyte Doolittle 1982, Hopp-Woods 1981).