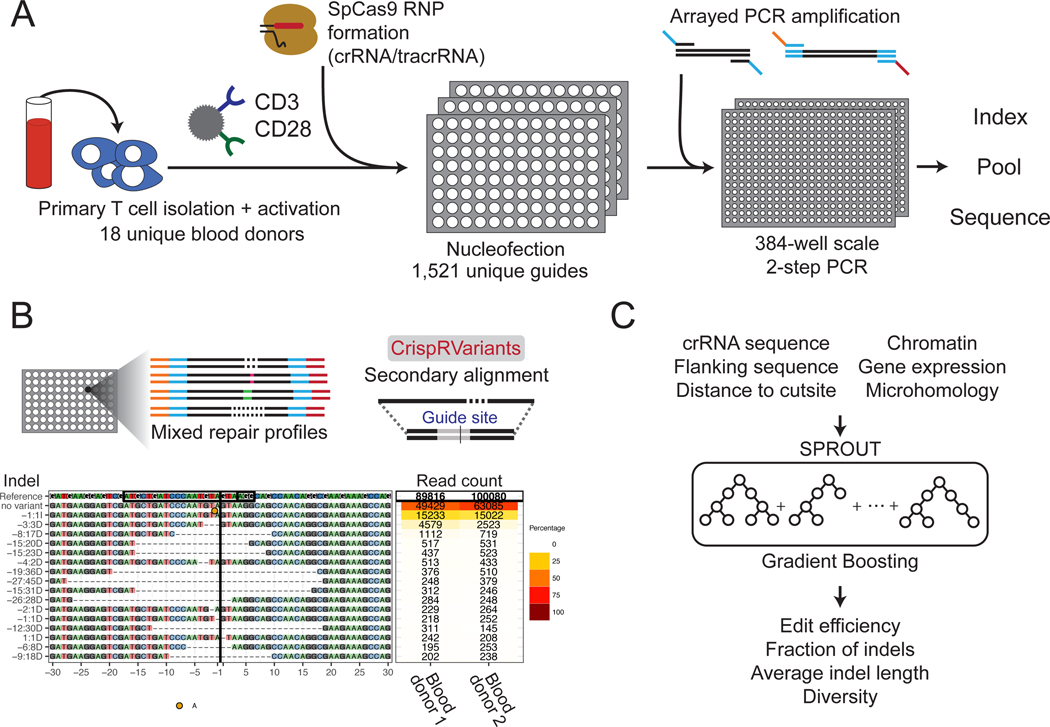
Figure 1.
Overview of the method. (A) Primary T cells were isolated, activated, and electroporated with Cas9/crRNA/tracrRNA RNPs in 96-well plates. After 6 days of expansion, genomic DNA was isolated from each well, amplified and sequenced. (B) The CrispRVariants R package4 was used to quantify each SpCas9 RNP knockout. An example alignment is plotted here, with quantification shown for two blood donors. Each site has this same unique plot, all of which can be found on figshare. (C) A gradient boosting machine learning algorithm was trained to predict multiple DNA repair outcomes given the guide RNA sequence, flanking nucleotides and additional features.