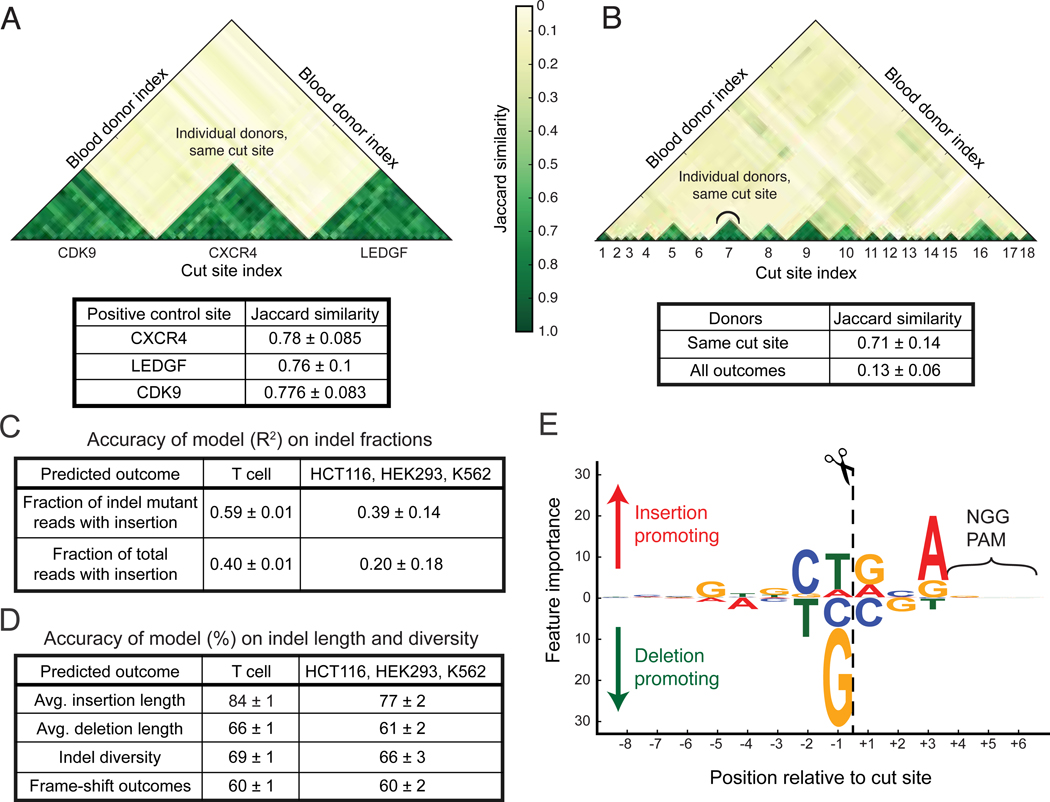
Figure 2.
SPROUT predicts DNA repair outcomes. (A) The DNA repair outcomes resulting from RNP activity in T cells derived from different blood donors were compared for control guides targeting CDK9, CXCR4, and LEDGF, analyzing the top 20 indels at each site. These guides were used in every blood donor. Jaccard similarity is calculated for each guide site across donors. (B) Jaccard similarity of DNA repair outcomes for 18 randomly chosen guides, again using the top 20 indels. Jaccard coefficients are plotted comparing outcomes from different guide RNAs and between blood donors. (C) The trained model was used to predict DNA repair indel fractions in a hold-out (un-seen) portion of the T cell dataset. The model was also evaluated on previously published data5 obtained from immortalized cell lines to test generalization performance for other cell types and experimental conditions. (D) Accuracy of the trained model in predicting the average insertion and deletion length, indel diversity and whether a target has high, medium or low fraction of frame-shift outcomes on both T cells and previously published data5. (E) The importance that SPROUT assigns to nucleotides at each position relative to the cut site. Larger text indicates that the presence of a particular nucleotide at a position has greater importance in determining the likelihood of insertion versus deletion. Bootstrap mean and standard deviation are shown in each table. This study assayed 1,656 genomic sites in T cells.