Abstract
Drug resistance and cellular adhesion are two key elements of both dissemination and prevalence of the human fungal pathogen Candida albicans. Smi1 belongs to a family of hub proteins conserved among the fungal kingdom whose functions in cellular signaling affect morphogenesis, cell wall synthesis and stress resistance. The data presented here indicate that C. albicans SMI1 is a functional homolog of Saccharomyces cerevisiae KNR4 and is involved in the regulation of cell wall synthesis. Expression of SMI1 in S. cerevisiae knr4Δ null mutants rescued their sensitivity to caspofungin and to heat stress. Deletion of SMI1 in C. albicans resulted in sensitivity to the cell-wall-perturbing compounds Calcofluor White and Caspofungin. Analysis of wild-type and mutant cells by Atomic Force Microscopy showed that the Young’s Modulus (stiffness) of the cell wall was reduced by 85% upon deletion of SMI1, while cell surface adhesion measured by Force Spectroscopy showed that the surface expression of adhesive molecules was also reduced in the mutant. Over-expression of SMI1, on the contrary, increased cell surface adhesion by 6-fold vs the control strain. Finally, Smi1-GFP localized as cytoplasmic patches and concentrated spots at the sites of new cell wall synthesis including the tips of growing hyphae, consistent with a role in cell wall regulation. Thus, Smi1 function appears to be conserved across fungi, including the yeast S. cerevisiae, the yeast and hyphal forms of C. albicans and the filamentous fungus Neurospora crassa.
Keywords: Fungal cell wall, Adhesion, Caspofungin, Candida albicans, Atomic Force Microscopy
Introduction
Fungal infections are responsible of the death of an estimated 1.5 million people per year worldwide. Yeasts of the Candida genus are the second most numerous agents of fungal infections, with a prominent contribution by Candida albicans, which causes over 400,000 cases of life-threatening systemic infections and 200,000 deaths per year (Brown et al., 2012). Only four classes of antifungal drugs are available for patient treatment and the emergence of resistance is becoming a serious concern. The fungistatic group of azoles and the more recently-developed fungicidal group of echinocandins constitute the two major classes of antifungals used to treat patients. Azoles block the biosynthesis of ergosterol – an essential sterol for fungal cell membranes – by targeting the cytochrome P450 14-α demethylase enzyme, Erg11, which catalyzes the conversion of lanosterol to ergosterol, thereby affecting membrane integrity and inhibiting fungal growth (Kathiravan et al., 2012, Vanden Bossche et al., 1995). On the other hand, echinocandins, a class of compounds developed between 2001 and 2006, target the catalytic subunit of the β-1,3 glucan synthase protein complex (Odds et al., 2003). Studies of the molecular mechanisms of resistance to these two classes of antifungal compounds (recently reviewed in (Scorzoni et al., 2017)) have revealed that there are three major mechanisms leading to resistance in C. albicans: overexpression of multidrug efflux pump-encoding genes, notably CDR1, CDR2 and MDR1 (Sanglard and Odds, 2002, Sanglard et al., 1995), amino acid substitutions in the target proteins (ex: Erg11, Fks1) and alteration in the levels of proteins involved in sensitivity to the drug (ex: Erg3, Erg11). In addition, the formation of fungal biofilms can also be considered as a form of antifungal resistance mechanism due to the ability of the biofilm extracellular matrix (ECM) to bind and entrap antifungal compounds, particularly azoles and amphotericin B (Desai et al., 2014, Taff et al., 2013, Vediyappan et al., 2010, Zarnowski et al., 2014).
In this context, alternative antifungal targets and/or ways to improve the fungicidal effect of existing antifungals are being sought. Such approaches notably involve targeting chaperones such as Hsp90 or components of stress signaling pathways, since these targets are more likely to simultaneously affect resistance to different classes of antifungals, morphogenesis mechanisms, cellular fitness and adaptation to changing environments. Key studies have been conducted in these areas by, for example, Brown and colleagues (Brown et al., 2010) and Cowen and coworkers (Singh et al., 2009). Works by this latter group established the complex connections between Pkc1, Hsp90 and calcineurin suggesting interesting new strategies to treat fungal infections (LaFayette et al., 2010). However, these cellular targets suffer from a major drawback in that they are conserved in mammalian host cells, which makes achieving fungal specificity a real challenge. Factors that regulate the pathogen’s cell wall therefore remain a strong target for new, fungus-specific, therapeutic approaches.
Here we describe the role of Smi1, a C. albicans protein homologous to the Saccharomyces cerevisiae hub protein, Knr4, which interacts physically with both the Slt2 MAP kinase and calcineurin, thus connecting the two primary signaling pathways involved in cell wall maintenance during stress: the cell wall integrity pathway (CWI) and the calcineurin pathway (Dagkessamanskaia et al., 2010, Martin-Yken et al., 2016). Although its precise molecular mode of action is currently unknown, it has been shown that two conserved serine residues, S200 and S203, phosphorylated in vivo, are essential for Krn4 function in signal transmission (Ficarro et al., 2002, Basmaji et al., 2006). Knr4 is required for resistance to cell wall stress induced by elevated temperature or by the presence of antifungal compounds, including caspofungin (Lesage et al., 2004, Markovich et al., 2004). Knr4 also plays a role in filamentous and pseudohyphal growth, mucin secretion and agar invasion (Birkaya et al., 2009). Similarly, GS1 protein, the homolog of Knr4 and Smi1 in the model filamentous fungus, Neurospora crassa, is also involved in the control of morphogenesis, caspofungin sensitivity and the synthesis of cell wall constituents, notably β-glucans (Enderlin and Selitrennikoff, 1994, Resheat-Eini et al., 2008, Seiler and Plamann, 2003).
The C. albicans genome encodes two homologs of KNR4: SMI1 and SMI1B. Previous studies have shown that deletion of SMI1 affects cell wall β-glucan synthesis, biofilm formation and biofilm extracellular matrix production, as well as biofilm-associated resistance to fluconazole (Nett et al., 2011). Global transcriptomic studies indicate that SMI1 expression is induced in hyphal and planktonic cells by the Cyr1 adenylate cyclase, a positive regulator of C. albicans hyphal morphogenesis, and is also biofilm-induced, while it is repressed by the Hap43 regulatory protein and caspofungin (Liu et al., 2005). Much less is known about SMI1B, although it appears to be the closest homolog of S. cerevisiae KNR4 according to phylogeny.
In this work we further characterize the function of SMI1 in C. albicans. We provide evidence that Smi1 is a functional homolog of S. cerevisiae Knr4, that its correct expression is critical for the regulation of fungal cell wall integrity and biophysical properties, and that the cellular localization of Smi1-GFP in yeast and hyphal cells is consistent with those observed for its counterparts, i.e. Knr4 in S. cerevisiae yeasts and GS1 in the hyphae of N. crassa.
Material and methods
Strains and growth media
The C. albicans and S. cerevisae strains used in this study are listed in Table 1. Depending on experimental conditions, yeast strains were grown in YPD (1% (W/V) yeast extract, 2% peptone, and 1% dextrose), YP (1% (W/V) yeast extract, 2% peptone) supplemented with 10% Fetal Bovine Serum (FBS), or SD (synthetic dextrose, 0.67% (W/V) yeast nitrogen base (YNB; Difco) with 2% glucose) supplemented as necessary with arginine, histidine or uridine (20 mg/l). Agar (2%) was used for growth on solid medium. Escherichia coli strains TOP10 (Invitrogen) or DH5α were used for DNA cloning and maintenance of the plasmid constructs.
Table 1.
Yeast strains used in this study.
| Strain name | Genotype | Reference or Source |
|---|---|---|
| Candida albicans strains: | ||
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434, arg4Δ::hisG/arg4Δ::hisG, his1Δ::hisG/his1Δ::hisG | Wilson et al. (1999) |
| CEC161 | Isogenic to BWP17 but arg4Δ::hisG/ARG4, his1Δ::hisG/HIS1 | Firon et al. (2007) |
|
BWP17 AHU (= CEC 369) |
Isogenic to CEC161 but ura3::limm434/URA3 | Moreno-Ruiz et al. (2009) |
| smi1 Δ/Δ | Isogenic to BWP17 but smi1Δ::HIS1/smi1Δ::ARG4, RPS1/rps1::CIp10 | This study |
| smi1 Δ/Δ+PTDH3SMI1 | Isogenic to smi1Δ/Δ but RPS1/rps1::CIp10-PTDH3-SMI1 | This study |
| SMI1-OE | Isogenic to CEC161 but RPS1/rps1::CIp10-PTDH3-SMI1 | This study |
| DAY185 | ura3::imm434/ura3::imm434 his1::hisG::HIS1/his1::hisG arg4::hisG::ARG4-URA3/arg4::hisG | Davis et al. (2000) |
| SMI1-GFP | DAY185 SMI1::SMI1-GFP-NAT | This study |
| Saccharomyces cerevisiae strains: | ||
| BY4741a | MAT a; his3Δ1 leu2Δ0; met15Δ0; ura3Δ0 | Brachmann et al. (1998) |
| knr4Δ | BY4741aYGR229c::KanMX4 | YKO Collection (Open Biosystems) |
| W303-2N | MAT a/α ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100 | Rodney Rothstein. |
| HM1315 | W303-2N YGR229c::KanMX4/YGR229c::KanMX4 | This study. |
Plasmid construction and generation of epitope-tagged or mutant strains
C. albicans SMI1 gene was PCR amplified with primers: Sense SMI1 New start and Antisense SMI1 (sequences in Supplementary material Table S1). The PCR product was then cloned in the S. cerevisiae expression vector YEplac195 PGK/CYC1 between the S. cerevisiae PGK1 promotor PPGK1 and the S. cerevisiae CYC1 terminator sequences (personal gift of Dr. J.L. Parrou, based on YEplac195 (Gietz and Sugino, 1988)), thus yielding the pSMI1 plasmid. Plasmid pKNR4 expressing the S. cerevisae KNR4 gene with its own promotor and terminator on a multicopy vector has been described previously (Martin et al., 1999).
S. cerevisiae cells were transformed using the lithium acetate method (Gietz and Woods, 2006). C. albicans cells were transformed using the lithium acetate protocol of (Walther and Wendland, 2003), followed by selection of transformants for uridine, arginine or histidine prototrophy when using the URA3, ARG4 or HIS1 markers, respectively.
Construction of C. albicans original smi1 Δ/Δ and smi1B Δ/Δ knock-out mutants used PCR-generated ARG4 and HIS1 disruption cassettes flanked by 120 base pairs of target homology region (primer sequences are provided in Supplementary material Table S1) as described by (Gola et al., 2003) and (Schaub et al., 2006). Independent transformants were produced and the gene replacements were verified by PCR on whole yeast cells as described previously (Gola et al., 2003, Schaub et al., 2006).
The SMI1 (C1_07870C_A) gene was amplified using primers: SMI1 Forward and SMI1 Reverse (sequences provided in Supplementary material Table S1). The resulting 1.8 kb PCR product was purified and inserted into the GTW sequences of pEntry (Gateway™ system, Invitrogen). Recombination of pEntry-SMI1 plasmid and CIp10-PTDH3-GTW plasmid (Chauvel et al., 2012) was performed also using the Gateway™ (system Invitrogen). CIp10-PTDH3-GTW vector is a derivative of plasmid CIp10 (Murad et al., 2000) that carries the sequence for integration at the RPS1 locus on C. albicans Chr1, the URA3 gene, and a Gateway™ cassette flanked by the attR sequences and preceded by the C. albicans PTDH3 constitutive promoter (Delgado et al., 2003). The resulting CIp10-PTDH3-SMI1 construct was then used to transform by genome integration through targeted homologous recombination at the genomic RPS10 locus the host strain BWP17, yielding SMI1-OE strain, as well as the smi1 Δ/Δ original mutant, yielding smi1 Δ/Δ+PTDH3 SMI1. To allow phenotype comparisons with the SMI1-OE strain, we used as control strain BWP17 AHU (Moreno-Ruiz et al., 2009) and the smi1 Δ/Δ original mutant was also transformed by the empty Clp10 vector, ensuring that all of these strains carry a functional URA3 allele (see Table 1 for full genotypes of the yeast strains used in this study).
GFP tagging of Smi1
Smi1 was C-terminally-tagged with GFP by amplifying GFP-NAT cassette from the pGFP-NAT1 plasmid (Milne et al., 2011) using primers gSmi1 _Fa and gSmi1_Ra containing 100 bp of flanking homology to the SMI1 terminator and the C-terminus of the SMI1 ORF (C1_07870C_A), respectively (primer sequences are provided in Supplementary material Table S1). Transformants were selected on YPD agar containing 300 µg/ml nourseothricin (Sigma). Integration of the GFP-NAT cassette was confirmed by PCR using primers sSmi1_F and sFP_R, which anneal to the chromosome outside the targeted region and within the cassette, respectively (primer sequences in Supplementary material Table S1).
Microscopy of Smi1-GFP
Yeast were grown for 2 h at 30 °C in YNB medium containing amino acids and (NH4)2SO4 (Sigma Aldrich). Morphogenesis was induced at 37 °C for 3 h in 20% foetal calf serum (FCS), 2% glucose. Cells were imaged on µ-slides (Ibidi, Martinsried, Germany). Images of Smi1-GFP localization were captured on an UltraVIEW® VoX spinning disk confocal microscope (Nikon, Surrey, UK), using a 488 nm laser. Multiple Z-stack images were acquired and Z-stack projections at maximum intensity were created using Volocity 6.3 software (Perkin Elmer).
Phenotypic sensitivity tests
Drop tests to evaluate the sensitivity of different strains and mutants to cell wall affecting drugs were performed as previously described (Ram et al., 1998) with minor modifications (Martin et al., 1999). Briefly, yeast cells were grown in liquid YPD to OD600 of 1 ± 0.1, then concentrated by centrifugation and resuspended in sterile water to an OD600 of 8. Serial dilutions of 1/1, 1/10, 1/100 and 1/1000 were then spotted on solid media containing either calcofluor white or Caspofungin at the indicated concentrations. Growth was scored and photographs taken after 48 h of incubation at 30 °C, or at 37 °C for testing the sensitivity to elevated temperature.
Atomic Force Microscopy (AFM)
Sample preparation for AFM experiments: Yeast cells were concentrated by centrifugation, washed two times in acetate buffer (18 mM CH3COONa, 1 mM CaCl2, 1 mM MnCl2, pH 5.2), resuspended in the same buffer, and immobilized on polydimethylsiloxane (PDMS) stamps prepared as described by (Dague et al., 2011, Formosa et al., 2014a). Briefly, freshly oxygen-activated microstructured PDMS stamps were covered with a total of 100 μl of cell suspension and allowed to stand for 15 min at room temperature. Yeast cells were then deposited into the stamps microstructures by convective (capillary) assembly.
AFM procedures: For imaging and force spectroscopy, we used an AFM Nanowizard III (JPK Instruments, Berlin, Germany). Force curves were then recorded in acetate buffer in quantitative-imaging mode (JPK Instruments, 2011, QITM mode-quantitative imaging with Nano-Wizard 3 AFM) (Chopinet et al., 2013, Formosa et al., 2014b, Smolyakov et al., 2016) with MLCT AUWH cantilevers (nominal spring constants: 0.01, 0.1, and 0.5 N/m). For imaging, cantilevers with a spring constant of 0.01 N/m were used. For force spectroscopy experiments, cantilevers with spring constants of 0.1 and 0.5 N/m were used. The maximal applied force was kept at 1 nN, the force curves lengh (Z-range) at 2 μm and the approach/retract speed at either 20 or 2 μm s−1 for both imaging and force spectroscopy. The spring constant of each cantilever was determined by the thermal-noise method (Hutter and Bechhoefer, 1993). For elasticity measurements, force maps of 32 by 32 or 64 by 64, hence either 1024 or 4096 force curves were recorded on an area of 1–4 μm2 on top of the cells, always avoiding any bud or budscar. The force-distance curves recorded were transformed into force-indentation curves by subtracting the cantilever deflection on a solid surface. The indentation curves were then fitted to the Hertz model (Hertz, 1881).
Results
Conservation of cellular function between S. cerevisiae Knr4 and C. albicans Smi1
The genome of the human fungal pathogen C. albicans contains two distinct homologs of the S. cerevisiae KNR4 gene, SMI1 (C1_07870C_A) and SMI1B (C3_05350C_A). Gene deletion mutants for each gene were generated and initial phenotypic analysis showed a strong phenotype (sensitivity to Calcofluor White (CFW) or SDS) for the smi1 Δ/Δ mutant but only a milder one for the smi1B Δ/Δ mutant (Supplementary material, Fig. S1). We therefore focused on the role of Smi1 in this study.
The SMI1 open reading frame was amplified and cloned into a S. cerevisiae expression vector, under the control of the strong and constitutive ADH1 promoter. The SMI1 coding sequence is 1863 bp and contains two CTG codons at positions 1717 and 1762, which have a 97% chance of translation as a Serine in C. albicans through non-canonical codon usage in this fungus, compared to a Leucine in S. cerevisiae (White et al., 1995). These two codons are located within the C-terminus of Smi1, which is less conserved than the central domain among members of the fungal Knr4/Smi1 super-family of proteins (See Supplementary material, Fig. S2). The C-terminus of Knr4 is largely unstructured and not directly necessary for protein function in S. cerevisiae (Dagkessamanskaia et al., 2010, Martin-Yken et al., 2016). Hence, we tested the ability of the C. albicans SMI1 gene, retaining its two ambiguous codons, to complement the cell wall-related phenotypes of S. cerevisiae knr4 null mutants. Our phenotypic screen included sensitivity to CFW, SDS, caspofungin and elevated temperature in haploid and diploid S. cerevisiae genetic backgrounds. CFW is a compound that binds to nascent chitin fibrils and has been used to identify fungal cell wall mutants (Ram et al., 1994). Expression of SMI1 in S. cerevisiae was able to complement these phenotypes (Fig. 1A and data not shown), including the growth defect at elevated temperature (Fig. 1B), a specific defect in this organism that is linked to cell cycle progression through START (Fishel et al., 1993, Martin-Yken et al., 2003). The function of the Smi1/Knr4 proteins thus appears to be conserved between the two species despite their phylogenetic distance and the differences between the two protein sequences, which share only 34% identity and 49% similarity (Fig. S2).
Fig. 1.
C. albicans SMI1 gene expression suppresses the cell wall associated phenotypes of S. cerevisiae knr4 Δ mutants. A) Transformed haploid control strain BY4741a and mutant strain knr4Δ with either empty plasmid YEplac195 PGK/CYC1, pSMI1 (Yeplac195 bearing C. albicans SMI1 gene under PPGK1) or pKNR4 bearing S. cerevisiae KNR4 gene, were grown in liquid SD medium lacking uracil at 30 °C to an OD600 of 1, and concentrated to OD600 8 ± 0. Serial dilutions of yeast cultures were spotted on YPD plates in the absence or presence of 150 ng caspofungin ml−1. Growth was scored after 2 days at 30 °C. B) Transformed diploid control strain W3032N and mutant strain HM1315 knr4 Δ/Δ with either empty YEplac195 PGK/CYC1 plasmid, pSMI1 bearing C. albicans SMI1 gene or pKNR4, were grown overnight in liquid SD medium lacking uracil at 30 °C and concentrated to OD600 8 ± 0. Serial dilutions of yeast cultures were spotted on YPD plates. Growth was scored after 2 days at 30 °C and 37 °C.
Deletion of SMI1 renders C. albicans sensitive to cell wall targeting drugs
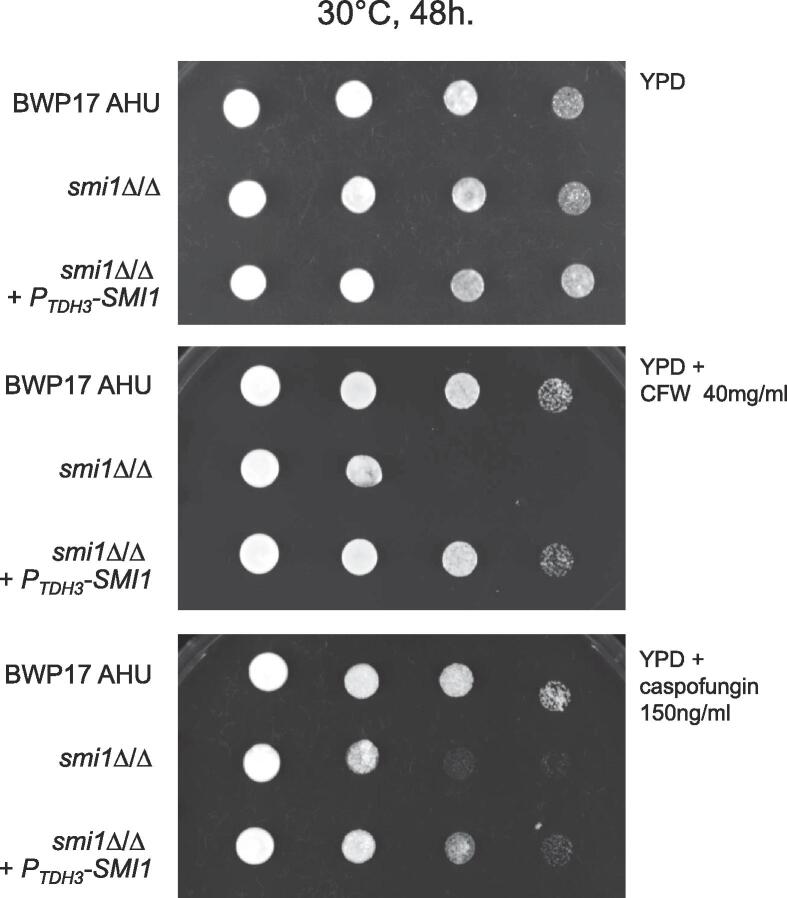
We further investigated the role of Smi1 in maintaining cell wall integrity using a medically-relevant β-glucan synthase-targeting drug, the echinocandin caspofungin. The influence of Smi1 on the sensitivity to CFW was also retested in parallel. Deletions of both alleles of SMI1 in the C. albicans BWP17 strain background (but bearing a functional URA3 allele) led to a marked increase in sensitivity to caspofungin and CFW at 30 °C (Fig. 2). These phenotypes are consistent with those observed for the S. cerevisiae knr4Δ mutant and the proposed role of these proteins in stress signaling pathways (See Discussion). Re-integration of the SMI1 open reading frame under the constitutive promoter PTDH3 in the smi1 Δ/Δ deletion mutant restored the wild-type phenotype (Fig. 2).
Fig. 2.
Calcofluor White and caspofungin sensitivity of the C. albicans smi1 Δ/Δ mutant. The control strain BWP17 AHU, the mutant strain smi1 Δ/Δ and the deletion mutant with SMI1 gene re-integrated smi1 Δ/Δ + PTDH3-SMI1 were grown in liquid YPD medium at 30 °C to an OD600 of 1, and concentrated to OD600 8 ± 0. Serial dilutions of yeast cultures were spotted on YPD plates in the absence or presence of 40 mg of CFW or 150 ng caspofungin ml−1. Growth was scored after 2 days at 30 °C.
Cell wall biophysics
Cell wall strength/elasticity
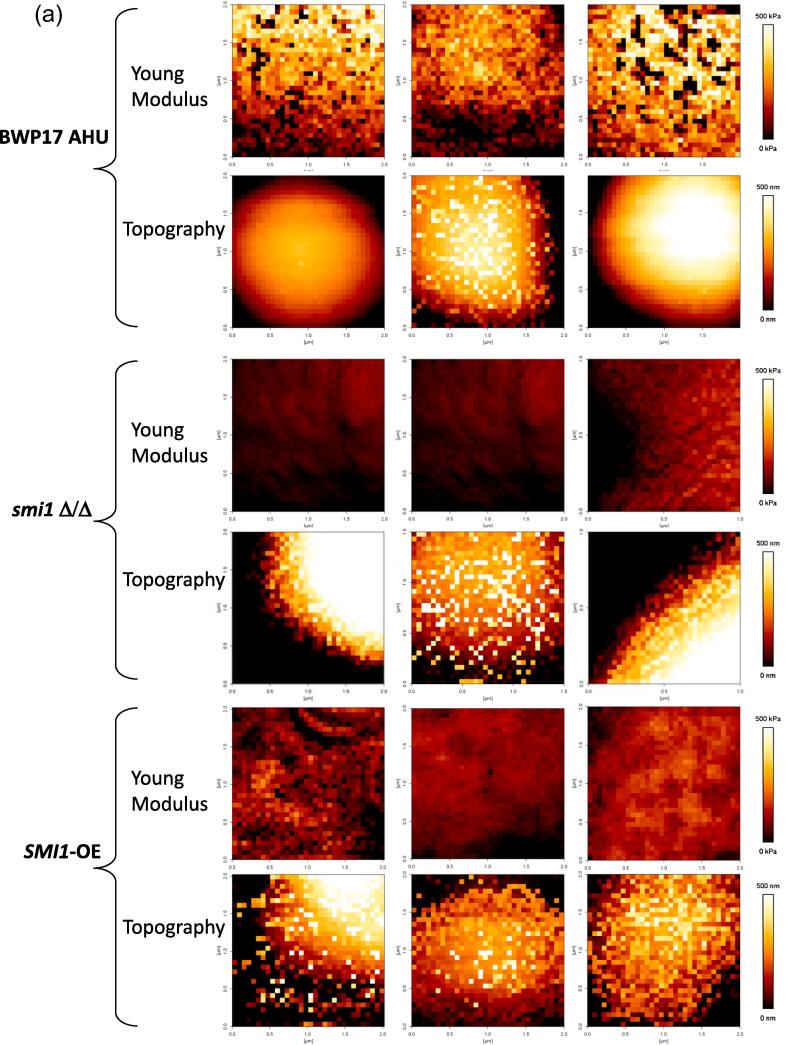
Atomic Force Microscopy (AFM) under liquid conditions can be used to investigate the nanomechnical properties of live wildtype and mutant cells (Dague et al., 2010, Ene et al., 2015, Formosa et al., 2013, Liu et al., 2015). Here we first measured the cell surface elasticity of three C. albicans strains: the control strain, BWP17 AHU, the homozygous deletion mutant smi1 Δ/Δ and the strain over-expressing the SMI1 from the PTDH3 promoter in the BWP17 genetic background, SMI1-OE. Using the Atomic Force Microscope in the Force Volume mode, we collected between 1024 and 4160 force curves per cell on a minimum of 12 cells from 4 independent cultures for each strain. The elasticity of the cells was quantified from these curves by calculating the Young’s Modulus (YM) as previously described (Dague et al., 2010). The Young’s Modulus represents the cell’s stiffness: the higher the YM, the stiffer the cell. In the control strain, BWP17 AHU, the mean value ± SEM of the YM was 782 ± 70 kPa (Fig. 3, Table 2), significantly higher than that of the homozygous deletion mutant (mean ± SEM = 93 ± 33 kPa). The YM of the over-expression strain (mean ± SEM = 298 ± 62 kPa) lay between the control strain and the deletion mutant, suggesting either a gene dosage effect for SMI1 expression or an effect of uncoupling the expression of this gene from the cell cycle. Fig. 3A shows maps of the recorded YM on square areas on the surfaces of three characteristic cells for each cell type, together with the corresponding topography maps. In this figure, the first line of maps (YM values of the control strain) is to compare with the third line (YM values of the homozygous deletion mutant) and the fifth line (YM values of the SMI1 over-expressing strain). In addition, all the YM values recorded are represented for each cell individually on Fig. 3B, to allow visualization of cell-to-cell variability. Finally, Fig. 3C shows selected representative individual force curves for the three strains. These curves represent the force encountered while approaching the AFM tip vertically toward the cell surface. Before the tip touches the cell it does not encounter any resistance and the curve is simply horizontal. When the tip touches the cell (contact point), the curve starts to bend. Moving the tip further down results in indenting into the cell surface, where distinct resistance levels can be met. Hence, the slope of the second part of each curve represents the cell surface resistance against the ongoing progression of the tip: the steeper the slope, the harder the surface. These results therefore demonstrate that the deletion of both alleles of SMI1 resulted in a reduction in cell wall stiffness by eight to ten folds, indicating that the cell wall integrity is compromised in this mutant.
Fig. 3.
A: Elasticity maps recorded on independent cells of BWP17 AHU, smi1 Δ/Δ and SMI1-OE strains. Maps of Young’s Moduli (YM = 1/Elasticity) measured by Atomic Force Microscopy on independent cells of control strain BWP17 AHU, smi1 Δ/Δ mutant and SMI1-OE strain. YM scales are shown (bright yellow: maximum at 500 kPa; dark red: minimum at 0.0 kPa). The corresponding topography map is presented below each elasticity map, also with scale (bright yellow: maximum at 500 nm; dark red: minimum at 0.0 nm). Analyzed areas cover squares of 1 × 1 to 2 × 2 µm2. B: Young’s Moduli of smi1 Δ/Δ mutant and SMI1-OE vs control strain BWP17 AHU. Atomic Force Microscope was used to collect over 12,300 force curves for each strain on the control strain BWP17 AHU, the smi1 Δ/Δ mutant and the SMI1-OE strain. The Young’s Moduli quantified from these curves are presented here as a dot on the mean YM value, with SEM for each cell. The bar represents the mean of the YM values with each SEM. Statistical analysis was done using the One-way ANOVA test, **** = p value < 0.0001. C: Representative Approach Force Curves of BWP17 AHU, smi1 Δ/Δ and SMI1-OE. Forces measured by AFM in nN as a function of the indentation (tip position) in nm, for the three strains. BWP17 AHU: red curves, smi1 Δ/Δ: black curves and SMI1-OE: blue. These force curves are obtained upon approaching the AFM tip towards the cell surface (horizontal part), touching the cell (contact point: where the curve starts to bend), further indenting into the cell surface and facing distinct resistance levels. The slope of the second part of each curve corresponds to the cell surface resistance against the tip progression.
Table 2.
Summary of Atomic Force Microscopy measurements for BWP17 AHU, smi1 Δ/Δ, and SMI1-OE strains.
| Cell Type | Young Modulus (kPa)a |
% of Adhesive Eventsb |
Mean Adhesion Force (pN)c | Adhesion Energy (=Area below the force curve) (J)d |
|---|---|---|---|---|
| BWP17 AHU | 782 (±70) | 46.1 | 127 (±33) | 1.77 × 10−17 |
| smi1 Δ/Δ | 93 (±33) | 19.4 | 70 (±16) | 0.26 × 10−17 |
| SMI1-OE | 298 (±62) | 62.9 | 712 (±102) | 15.82 × 10−17 |
aMean values with standard deviation of Young’s Moduli calculated from force curves obtained as described above (3.1).
bPercentage of adhesive events measured by AFM, calculated from at least 12,000 force curves for each cell type, with a threshold level for the definition of an adhesive event as 50pN on the retraction curve.
cMean values of Adhesion forces for each cell type, calculated from adhesive force curves obtained as described above.
dMean values of the Adhesion Energy for each strain, calculated from the area below the force curves presenting an adhesion event.
Adhesion
Another cell surface feature that can be easily and precisely measured by AFM is the ability to adhere to surfaces, using Single Molecule Force Spectroscopy (Axner et al., 2010, Benoit et al., 2000, Formosa et al., 2014a, Hinterdorfer et al., 1996, Neuman and Nagy, 2008). Here, adhesion between the cell surface and the AFM bare tips, constituted of Si3N4, was measured by scanning areas of 1 µm2 on the top of individual yeast cells. We recorded force curves whose retraction values were used to generate adhesion maps where the intensity of each pixel corresponds to the force required to dissociate the AFM tip from the sample, i.e. the adhesion force, expressed in picoNewtons pN (Fig. 4). These data, represented as two-dimensional matrixes, show that the adhesion between the probe and the cell surface of the smi1 Δ/Δ mutant was minimal compared to the control strain, while the cells of the SMI1 over-expression strain displayed a marked increase in their surface adhesion. Cellular adhesion was also evaluated by three quantitative parameters: the mean adhesion force, the specific energy of each adhesion event observed, and the overall frequency of these adhesive events among all the recorded force curves. In order to quantify these values, we defined as an adhesive event any force curve showing an adhesion force above 50pN. Using this threshold level, we calculated the percentage of adhesive events for each cell type and measured the area situated below the retraction curves, which represent the adhesion energy of the event (Table 2). These values indicated that adhesive events were encountered more frequently at the surface of the over-expressing strain SMI1-OE (63% of the recorded 21,500 force curves) than on the control strain (46% of over 13,500 force curves), and less frequently on the deletion mutant surface (19% of over 12,300 force curves). The mean adhesion force measured for the control strain was 127 ± 33 pN, calculated from 6200 adhesive force curves. This is to be compared with a mean ± SEM force of 70 ± 16 pN (barely above threshold) for 2300 force curves for the smi1 Δ/Δ mutant. For the SMI1-OE-strain, adhesive events were more frequent (63%) and they were also much stronger, with forces measured up to 2176 pN with a mean value ± SEM of 712 ± 102 pN, calculated over 13,000 adhesive force curves. The specific energy of these adhesive events also differed; with adhesion energies for the over-expressing strain an order of magnitude stronger than that for the control strain, while they were approximately seven times lower on the surface of the deletion mutant. Hence, the homozygous deletion of SMI1 gene abrogates almost entirely the ability of the mutant cell to adhere using the chemistry described here, while SMI1 over-expression leads to a highly adhesive phenotype.
Fig. 4.
Adhesion Maps recorded on the cellular surfaces of smi Δ/Δ mutant and SMI1-OE vs control strain. Adhesion force measurements were performed by Single Molecule Force Spectroscopy on C. albicans cells of the control strain BWP17 AHU, the smi1 Δ/Δ mutant and the SMI1-OE strain over-expressing the SMI1. The adhesion maps presented have been recorded on three independent and representative C. albicans cells for each cell type. Each analyzed area covers 1 × 1 to 2 × 2 µm2. Adhesion scales are shown and read as follows: bright yellow = maximum adhesion force at 2 nN; dark red = minimum at 0.0 nN.
Cellular localization
GS-1, the homolog of Smi1 and Knr4 in the model filamentous fungus, N. crassa, localizes at the growing tip of hyphae as a sphere positioned at the “Spitzenkörper” (Verdin et al., 2009). The Spitzenkörper (or apical body) is a fungal structure specific to true hyphae, located at the hyphal tip. It is composed of the secretory vesicles that are required for continuous polarized growth (Girbardt, 1957, Harris et al., 2005). GS-1-GFP and Knr4-GFP have been imaged at the tip of N. crassa hyphae (Riquelme et al., 2011; Sánchez-León et al., 2011; Verdin et al., 2009) and at the tip of elongated shmoos in S. cerevisiae, respectively ((Martin-Yken, 2012) and our unpublished data). To test whether the C. albicans homolog would be similarly positioned, a GFP-tag was integrated at the C-terminus of the Smi1 protein at its chromosomal locus and its cellular localization was visualized by confocal fluorescent microscopy in yeast and hyphal cells. This Smi1-GFP fusion protein is functional, as attested by its ability to complement the caspofungin and CFW hypersensitivity phenotypes of the smi1 Δ/Δ mutant (not shown).
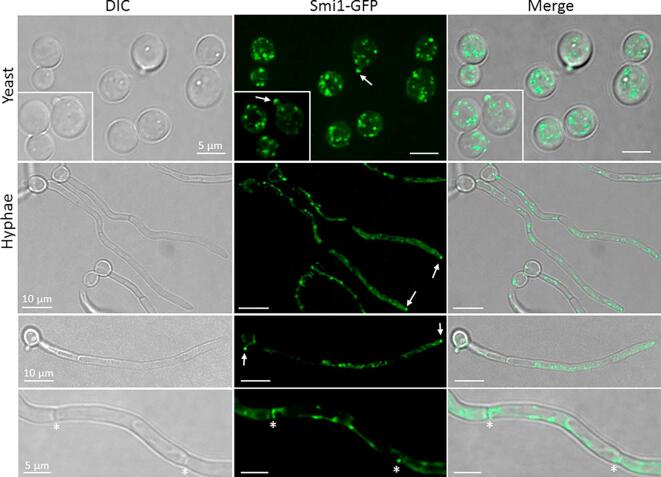
In yeast cells, Smi1-GFP appeared both as punctate patches in the cytoplasm and localized transiently to nascent buds (Fig. 5). This localization is similar to that reported for Knr4 in S. cerevisiae (Dagkessamanskaia et al., 2010, Martin et al., 1999). Punctate patches and a more diffuse cytoplasmic distribution were observed in C. albicans hyphae (Fig. 5), but, unlike in yeast, Smi1-GFP signal was consistently retained as a bright spot at the growing hyphal tip throughout the cell cycle, reminiscent of N. crassa GS-1 localization at the tip of growing hyphae (Verdin et al., 2009). Smi1-GFP was occasionally observed at hyphal septa as a dim signal, but this presence did not reflect specific stages of the cell cycle. Hence, Smi1 in C. albicans appears to associate with intracellular organelles and localizes to sites of new cell wall growth, a pattern which reflects those observed for homologs of Smi1 in S. cerevisae in yeast cells and the hyphae of N. crassa.
Fig. 5.
Smi1-GFP localizes as patches concentrated to apical growth sites in yeasts and hyphae. Cells were grown on Ibidi µ-slides in YNB medium at 30 °C for 2 h (yeast) and 20% FBS, 2% glucose at 37 °C for 3 h (hyphae). Smi1-GFP localized transiently to emerging bud tips in yeasts (arrows) and to septa in hyphae (asterisks) but was maintained consistently at hyphal/branch tips (arrows). Punctate fluorescence patches were also observed throughout yeast and hyphal cells. Images are maximum projections of individual z-stacks.
Discussion
Our results indicate that there is significant conservation of Knr4/Smi1 function in cell wall regulation between S. cerevisiae and C. albicans and demonstrate the role of Smi1 in tolerance to caspofungin, regulation of cell wall integrity and cell surface adhesion properties of this major human fungal pathogen. These features suggest that Smi1 might be relevant as a new drug target for combination therapies. Previous work by Nett and colleagues identified a role for Smi1 in the production of extracellular matrix during biofilm formation and hence the associated resistance to Fluconazole (Nett et al., 2011). Their results indicated that these effects were linked to the cell wall integrity pathway but were in fact regulated by Smi1 independently of the CWI pathway, suggesting a control pathway for Smi1 distinct from that of the PKC pathway. Lafayette and colleagues dissected the mechanisms through which PKC regulates resistance to both azoles and echinocandins in the two yeast models, C. albicans and S. cerevisiae (LaFayette et al., 2010). They showed that, in C. albicans, Pkc1 and calcineurin signaling pathways independently regulate antifungal resistance via a common unknown target, which they designed as “X” (see (LaFayette et al., 2010), Fig. 9B thereof). Considering the knowledge accumulated on Knr4 in budding yeast together with the data obtained for C. albicans and presented here, we propose that Smi1 is a candidate for this previously unidentified “X”, a common target of the Pkc1 and calcineurin signaling pathways. The marked reduction in cell-wall stiffness of the smi1 Δ/Δ cell wall, as indicated by its Young’s Modulus (Fig. 3, Table 2), compared to the milder phenotype observed for the S. cerevisiae knr4Δ mutant (Dague et al., 2010), suggests a more central role for Smi1 in the cell-wall integrity signaling network in C. albicans than that of Knr4 in the baker’s yeast, which is in agreement with the hypothesis of LaFayette and colleagues (LaFayette et al., 2010). The fact that the homozygous deletion mutant smi1 Δ/Δ is so strongly affected, despite the presence in this strain of both functional alleles of SMIB, argues for a major role for the SMI1 gene, at least in the conditions tested (30 °C, liquid rich medium, yeast form of C. albicans cells).
Our results show that homozygous deletion of SMI1 leads to an increase in the sensitivity of C. albicans to both CFW and caspofungin at 30 °C. Caspofungin tolerance has been reported in C. albicans mutants that have elevated cell-wall chitin (Perlin, 2015, Plaine et al., 2008, Walker et al., 2013, Yang et al., 2017), yet others report that elevated chitin can induce hypersensitivity to CFW (Elorza et al., 1983; Roncero and Durán, 1985). However, this dual sensitivity is consistent with the phenotype observed for the S. cerevisiae knr4 Δ mutant (Lesage et al., 2004, Martin et al., 1999). Our cell wall stiffness measurements indicated that organization of the cell wall is significantly modified in the deletion mutant smi1 Δ/Δ, consistent with a possible upstream role of Smi1 in cell-wall regulatory pathways. Early results obtained in bakers’ yeast established a role for Knr4 in the transcriptional control of all S. cerevisiae chitin synthase genes (Martin et al., 1999), so the role of Smi1 in C. albicans may also include control of expression levels of cell wall biosynthesis genes. The complex interplay of cellular signaling pathways controlling C. albicans susceptibility to echinocandins has been described by Munro and colleagues (Walker et al., 2010), and it now seems possible that Smi1 is one piece of this puzzle. Indeed, a previous study investigating the effects of caspofungin on the cell walls of S. cerevisiae and C. albicans by AFM revealed that treatment with this echinocandin increased C. albicans cell wall stiffness and at the same time enhanced cell surface adhesion (Formosa et al., 2013). Although the high level of sensitivity to caspofungin of the C. albicans smi1Δ/Δ mutant meant that it was not possible to test this in this fungus, we speculate that Smi1 is a key player in the cellular response to caspofungin in C. albicans.
In S. cerevisiae, Knr4 is proposed to link the Ca2+/calcineurin/Crz1 signaling pathway with the Slt2/Mpk1 cell-wall integrity pathway. The cellular localization observed for Smi1-GFP in C. albicans yeasts and hyphae, as cytoplasmic patches and concentrated spots at the polarized growth sites, is consistent with the localizations reported for the MAP kinase Slt2 and calmodulin, the activator of calcineurin in S. cerevisiae (Brockerhoff and Davis, 1992, van Drogen and Peter, 2002). Therefore, Smi1 in C. albicans could act as a similar link between these two signaling pathways, giving it a central role in cell-wall integrity signaling. In the model filamentous fungus, N. crassa, GS1, the homolog of Smi1, localizes at the Spitzenkörper within the hyphal tip (Verdin et al., 2009; Sánchez-León et al., 2011). This localization is conserved in another ascomycete filamentous fungus, A. nidulans (also called Emericella nidulans) (Schultzhaus et al., 2015). The results presented here indicate that Smi1 also localizes in or around the Spitzenkörper of C. albicans hyphae in a similar manner to that observed for GS-1 in N. crassa. Given the cell wall related phenotypes reported for GS-1 mutants of N. crassa (Enderlin and Selitrennikoff, 1994, Resheat-Eini et al., 2008, Seiler and Plamann, 2003), the function of these proteins appears to be not only conserved between C. albicans and S. cerevisiae, but also to some extent in filamentous fungi.
Finally, the role of Smi1 in the control of cell wall synthesis, cellular adhesion and drug resistance is relevant to the search for new antifungal targets. An advantage of Smi1 as a drug target over Hsp90, calcineurin, Pkc1 or other MAP kinases is the specificity of the Knr4/Smi1 superfamily of proteins to the Fungal Kingdom as it is absent from host cells (Martin-Yken et al., 2016). In addition, since this protein family is conserved among fungi, including other fungal pathogens of mammals (C. glabrata and Aspergillus species notably) and also plants (ex: Magnaporthe grisea), developing drugs that target Smi1 might lead to broader antifungal applications in domains such as agriculture.
Acknowledgments
Acknowledgements
Dr. Jean-Luc Parrou (LISBP, Toulouse) for vector YEplac195 PGK/CYC1 and Gregory Da Costa (INRA, Jouy-en-Josas) for technical help.
SZ is an Institut Pasteur International Network Affiliate Program Fellow (Institut Pasteur de Tunis, Institut Pasteur, Paris) and has also been supported by grants from the European Commission (FinSysB PITN-GA-2008-214004), the Agence Nationale de la Recherche (KANJI, ANR-08-MIE-033-01) and the French Government’s Investissement d’Avenir Program (Institut de Recherche Technologique BIOASTER, ANR-10-AIRT-03) to Chd’E.
HMY acknowledges the FEBS society and the organizers of the Human Fungal Pathogen Schools 2015 and 2017 for a great introduction to C. albicans and other human fungal pathogens, as well as Formation permanente of the Centre INRA Toulouse Occitanie for financing her attendance to these researcher schools.
HMY is forever grateful to Frans M. Klis for his kindness throughout the years and his great advice to “Start working on C. albicans”.
Declarations of interest
None.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.tcsw.2018.10.002.
Appendix A. Supplementary data
References
- Axner O., Björnham O., Castelain M., Koutris E., Schedin S., Fällman E., Andersson M. DIVA. Springer Verlag; 2010. Unraveling the Secrets of Bacterial Adhesion Organelles Using Single Molecule Force Spectroscopy; pp. 337–362. [Google Scholar]
- Basmaji F., Martin-Yken H., Durand F., Dagkessamanskaia A., Pichereaux C., Rossignol M., Francois J. The “interactome” of the Knr4/Smi1, a protein implicated in coordinating cell wall synthesis with bud emergence in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2006;275:217–230. doi: 10.1007/s00438-005-0082-8. [DOI] [PubMed] [Google Scholar]
- Benoit M., Gabriel D., Gerisch G., Gaub H.E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- Birkaya B., Maddi A., Joshi J., Free S.J., Cullen P.J. Role of the cell wall integrity and filamentous growth mitogen-activated protein kinase pathways in cell wall remodeling during filamentous growth. Eukaryot. Cell. 2009;8:1118–1133. doi: 10.1128/EC.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast (Chichester Engl.) 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brockerhoff S.E., Davis T.N. Calmodulin concentrates at regions of cell growth in Saccharomyces cerevisiae. J. Cell Biol. 1992;118:619–629. doi: 10.1083/jcb.118.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J.P., Leach M.D., Nicholls S. The relevance of heat shock regulation in fungal pathogens of humans. Virulence. 2010;1:330–332. doi: 10.4161/viru.1.4.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004404. 165rv13-165rv13. [DOI] [PubMed] [Google Scholar]
- Chauvel M., Nesseir A., Cabral V., Znaidi S., Goyard S., Bachellier-Bassi S., Firon A., Legrand M., Diogo D., Naulleau C. A versatile overexpression strategy in the pathogenic yeast candida albicans: identification of regulators of morphogenesis and fitness. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopinet L., Formosa C., Rols M.P., Duval R.E., Dague E. Imaging living cells surface and quantifying its properties at high resolution using AFM in QI™ mode. Micron Oxf. Engl. 2013;1993(48):26–33. doi: 10.1016/j.micron.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Dagkessamanskaia A., El Azzouzi K., Kikuchi Y., Timmers T., Ohya Y., Francois J.M., Martin-Yken H. Knr4 N-terminal domain controls its localization and function during sexual differentiation and vegetative growth. Yeast. 2010;27:563–574. doi: 10.1002/yea.1804. [DOI] [PubMed] [Google Scholar]
- Dague E., Bitar R., Ranchon H., Durand F., Yken H.M., Francois J.M. An atomic force microscopy analysis of yeast mutants defective in cell wall architecture. Yeast. 2010;27:673–684. doi: 10.1002/yea.1801. [DOI] [PubMed] [Google Scholar]
- Dague E., Jauvert E., Laplatine L., Viallet B., Thibault C., Ressier L. Assembly of live micro-organisms on microstructured PDMS stamps by convective/capillary deposition for AFM bio-experiments. Nanotechnology. 2011;22 doi: 10.1088/0957-4484/22/39/395102. [DOI] [PubMed] [Google Scholar]
- Davis D., Edwards J.E., Mitchell A.P., Ibrahim A.S. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M.L., Gil M.L., Gozalbo D. Starvation and temperature upshift cause an increase in the enzymatically active cell wall-associated glyceraldehyde-3-phosphate dehydrogenase protein in yeast. FEMS Yeast Res. 2003;4:297–303. doi: 10.1016/S1567-1356(03)00159-4. [DOI] [PubMed] [Google Scholar]
- Desai J.V., Mitchell A.P., Andes D.R. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb. Perspect. Med. 2014;4 doi: 10.1101/cshperspect.a019729. a019729 a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F., Peter M. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 2002;12:1698–1703. doi: 10.1016/s0960-9822(02)01186-7. [DOI] [PubMed] [Google Scholar]
- Enderlin C.S., Selitrennikoff C.P. Cloning and characterization of a Neurospora crassa gene required for (1,3) beta-glucan synthase activity and cell wall formation. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9500–9504. doi: 10.1073/pnas.91.20.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene I.V., Walker L.A., Schiavone M., Lee K.K., Martin-Yken H., Dague E., Gow N.A.R., Munro C.A., Brown A.J.P. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio. 2015;6:e00986–15. doi: 10.1128/mBio.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro S.B., McCleland M.L., Stukenberg P.T., Burke D.J., Ross M.M., Shabanowitz J., Hunt D.F., White F.M. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Firon A., Aubert S., Iraqui I., Guadagnini S., Goyard S., Prévost M.-C., Janbon G., d’Enfert C. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol. Microbiol. 2007;66:1256–1275. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- Fishel B.R., Sperry A.O., Garrard W.T. Yeast calmodulin and a conserved nuclear protein participate in the in vivo binding of a matrix association region. Proc Natl Acad Sci USA. 1993;90:5623–5627. doi: 10.1073/pnas.90.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa C., Schiavone M., Martin-Yken H., François J.M., Duval R.E., Dague E. Nanoscale effects of caspofungin against two yeast species, Saccharomyces cerevisiae and Candida albicans. Antimicrob. Agents Chemother. 2013;57:3498–3506. doi: 10.1128/AAC.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa C., Pillet F., Schiavone M., Duval R.E., Ressier L., Dague E. Generation of living cell arrays for atomic force microscopy studies. Nat. Protoc. 2014;10:199–204. doi: 10.1038/nprot.2015.004. [DOI] [PubMed] [Google Scholar]
- Formosa C., Schiavone M., Boisrame A., Richard M.L., Duval R.E., Dague E. Multiparametric imaging of adhesive nanodomains at the surface of Candida albicans by atomic force microscopystudies. Nanomed. Nanotechnol. Biol. Med. 2014 doi: 10.1016/j.nano.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. Yeast Protocol. Humana Press; Totowa, NJ: 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method; pp. 107–120. [Google Scholar]
- Girbardt M. Der Spitzenkörper von Polystictus versicolor (L.) Planta. 1957;50:47–59. [Google Scholar]
- Gola S., Martin R., Walther A., Dünkler A., Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- Harris S.D., Read N.D., Roberson R.W., Shaw B., Seiler S., Plamann M., Momany M. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell. 2005;4:225–229. doi: 10.1128/EC.4.2.225-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz H. Ueber die Berührung fester elastischer Körper. J Für Reine Angew Math. 1881:156–171. [Google Scholar]
- Hinterdorfer P., Baumgartner W., Gruber H.J., Schilcher K., Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- Kathiravan M.K., Salake A.B., Chothe A.S., Dudhe P.B., Watode R.P., Mukta M.S., Gadhwe S. The biology and chemistry of antifungal agents: a review. Bioorg. Med. Chem. 2012;20:5678–5698. doi: 10.1016/j.bmc.2012.04.045. [DOI] [PubMed] [Google Scholar]
- LaFayette S.L., Collins C., Zaas A.K., Schell W.A., Betancourt-Quiroz M., Gunatilaka A.A.L., Perfect J.R., Cowen L.E. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Sdicu A.-M., Menard P., Shapiro J., Hussein S., Bussey H. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics. 2004;167:35–49. doi: 10.1534/genetics.167.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Formosa C., Dagkessamanskaia, Dague E., Francois J.M., Martin-Yken Combining Atomic Force Microscopy and genetics to investigate the role of Knr4 in Saccharomyces cerevisiae sensitivity to K9 Killer toxin. Lett. Appl. NanoBiosci. 2015;4:306–315. [Google Scholar]
- Liu T.T., Lee R.E.B., Barker K.S., Lee R.E., Wei L., Homayouni R., Rogers P.D. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 2005;49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovich S., Yekutiel A., Shalit I., Shadkchan Y., Osherov N. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2004;48:3871–3876. doi: 10.1128/AAC.48.10.3871-3876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H., Dagkessamanskaia A., Satchanska G., Dallies N., François J. KNR4, a suppressor of Saccharomyces cerevisiae cwh mutants, is involved in the transcriptional control of chitin synthase genes. Microbiol. Read. Engl. 1999;145(Pt 1):249–258. doi: 10.1099/13500872-145-1-249. [DOI] [PubMed] [Google Scholar]
- Martin-Yken H. Primosten; Croatia: 2012. Knr4/Smi1 family: conserved fungal chaperones of puzzling origin. [Google Scholar]
- Martin-Yken H., Dagkessamanskaia A., Basmaji F., Lagorce A., Francois J. The interaction of Slt2 MAP kinase with Knr4 is necessary for signalling through the cell wall integrity pathway in Saccharomyces cerevisiae. Mol. Microbiol. 2003;49:23–35. doi: 10.1046/j.1365-2958.2003.03541.x. [DOI] [PubMed] [Google Scholar]
- Martin-Yken H., François J.M., Zerbib D. Knr4: a disordered hub protein at the heart of fungal cell wall signalling. Cell Microbiol. 2016;18:1217–1227. doi: 10.1111/cmi.12618. [DOI] [PubMed] [Google Scholar]
- Milne S.W., Cheetham J., Lloyd D., Aves S., Bates S. Cassettes for PCR-mediated gene tagging in Candida albicans utilizing nourseothricin resistance. Yeast Chichester Engl. 2011;28:833–841. doi: 10.1002/yea.1910. [DOI] [PubMed] [Google Scholar]
- Moreno-Ruiz E., Ortu G., de Groot P.W.J., Cottier F., Loussert C., Prévost M.-C., de Koster C., Klis F.M., Goyard S., d’Enfert C. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiol. Read. Engl. 2009;155:2004–2020. doi: 10.1099/mic.0.028902-0. [DOI] [PubMed] [Google Scholar]
- Murad A.M.A., Lee P.R., Broadbent I.D., Barelle C.J., Brown A.J.P. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Nett J.E., Sanchez H., Cain M.T., Ross K.M., Andes D.R. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot. Cell. 2011;10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F.C., Brown A.J.P., Gow N.A.R. Antifungal agents: mechanisms of action. Trends Microbiol. 2003;11:272–279. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- Perlin D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015;1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaine A., Walker L., Da Costa G., Mora-Montes H.M., McKinnon A., Gow N.A.R., Gaillardin C., Munro C.A., Richard M.L. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008;45:1404–1414. doi: 10.1016/j.fgb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram A.F., Kapteyn J.C., Montijn R.C., Caro L.H., Douwes J.E., Baginsky W., Mazur P., van den Ende H., Klis F.M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of beta1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resheat-Eini Z., Zelter A., Gorovits R., Read N., Yarden O. Neurospora crassa colonial temperature sensitive 2, 4 and 5 (cot-2, cot-4 and cot-5) genes encode regulatory and structural proteins required for hyphal elongation and branching. Fungal Genet. Rep. 2008;55:32–36. [Google Scholar]
- Sanglard D., Odds F.C. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2002;2:73–85. doi: 10.1016/s1473-3099(02)00181-0. [DOI] [PubMed] [Google Scholar]
- Sanglard D., Kuchler K., Ischer F., Pagani J.L., Monod M., Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub Y., Dünkler A., Walther A., Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J. Basic Microbiol. 2006:416–429. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- Scorzoni L., de Paula e Silva A.C.A., Marcos C.M., Assato P.A., de Melo W.C.M.A., de Oliveira H.C., Costa-Orlandi C.B., Mendes-Giannini M.J.S., Fusco-Almeida A.M. Antifungal therapy: new advances in the understanding and treatment of mycosis. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Plamann M. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell. 2003;14:4352–4364. doi: 10.1091/mbc.E02-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolyakov G., Formosa-Dague C., Severac C., Duval R.E., Dague E. High speed indentation measures by FV, QI and QNM introduce a new understanding of bionanomechanical experiments. Micron. 2016;85:8–14. doi: 10.1016/j.micron.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Taff H.T., Mitchell K.F., Edward J.A., Andes D.R. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013:8. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bossche H., Koymans L., Moereels H. P450 inhibitors of use in medical treatment: focus on mechanisms of action. Pharmacol. Ther. 1995;67:79–100. doi: 10.1016/0163-7258(95)00011-5. [DOI] [PubMed] [Google Scholar]
- Vediyappan G., Rossignol T., d’Enfert C. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob. Agents Chemother. 2010;54:2096–2111. doi: 10.1128/AAC.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin J., Bartnicki-Garcia S., Riquelme M. Functional stratification of the Spitzenkorper of Neurospora crassa. Mol. Microbiol. 2009;74:1044–1053. doi: 10.1111/j.1365-2958.2009.06917.x. [DOI] [PubMed] [Google Scholar]
- Walker L.A., Gow N.A.R., Munro C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010;47:117–126. doi: 10.1016/j.fgb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Gow N.A.R., Munro C.A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 2013;57:146–154. doi: 10.1128/AAC.01486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A., Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 2003;42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- White T.C., Andrews L.E., Maltby D., Agabian N. The “universal” leucine codon CTG in the secreted aspartyl proteinase 1 (SAP1) gene of Candida albicans encodes a serine in vivo. J. Bacteriol. 1995;177:2953–2955. doi: 10.1128/jb.177.10.2953-2955.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.B., Davis D., Mitchell A.P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Zhang L., Wakabayashi H., Myers J., Jiang Y., Cao Y., Jimenez-Ortigosa C., Perlin D.S., Rustchenko E. Tolerance to caspofungin in Candida albicans is associated with at least three distinctive mechanisms that govern expression of FKS genes and cell wall remodeling. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski R., Westler W.M., Lacmbouh G.A., Marita J.M., Bothe J.R., Bernhardt J., Lounes-Hadj Sahraoui A., Fontaine J., Sanchez H., Hatfield R.D. Novel entries in a fungal biofilm matrix encyclopedia. mBio. 2014;5 doi: 10.1128/mBio.01333-14. e01333–14-e01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.