Fig 2. Identifying ‘PPI genes’ using nearest neighbor GWAS hits.
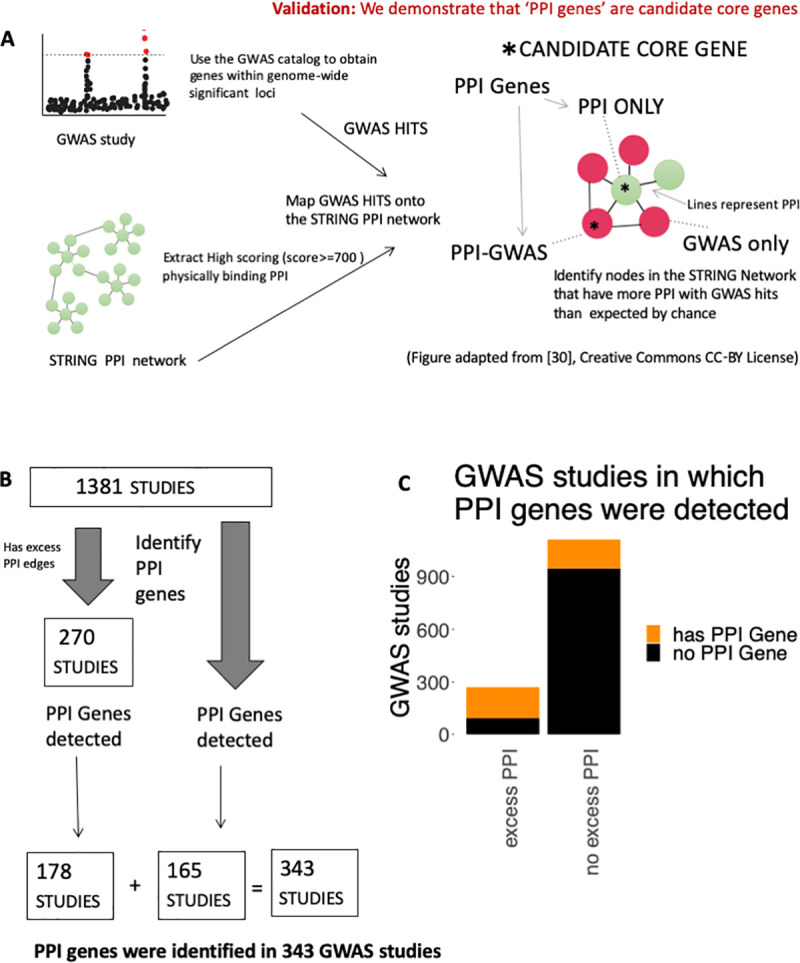
(A) This figure was adapted from [30] (Creative Commons CC-BY license), it illustrates our method for detecting PPI genes. This method involves putting GWAS hits from the GWAS catalog onto the STRING PPI network, and identifying neighboring nodes that have excess PPI with the GWAS hits. We also define the following terms. ‘GWAS’: Genome-wide Association Study, ‘GWAS HIT’ Gene within genome-wide significant loci (p < 5x10-8), ‘PPI’: Protein-Protein Interaction. ‘CORE GENE’: Gene with direct roles in disease biology ‘CANDIDATE CORE GENE’: Gene identified by our method that is likely to be a core gene. ‘PPI Gene’: PPI only or PPI-GWAS gene, ‘GWAS ONLY’: GWAS hit that is not a PPI gene. ‘PPI-GWAS’: PPI gene that is also a GWAS hit. ‘PPI only’: PPI gene that is not a GWAS hit. ‘All’: all 11,049 nodes within the STRING network. (B) Schematic demonstrating how we detected PPI genes in 343 Individual GWAS studies (178 of these studies had excess PPI between GWAS hits, while 165 studies did not show excess PPI among GWAS hits). (C) Proportion of studies with excess PPI that we also detect PPI genes in. Out of 270 studies in which we detect excess PPI amongst GWAS hits, we detect PPI genes in 178 studies. Amongst the remaining 1,111 GWAS studies, we detect PPI genes in 165 studies.
