Figure 2. Heatmap of copy number variations in 11 Ghanaian tumors.
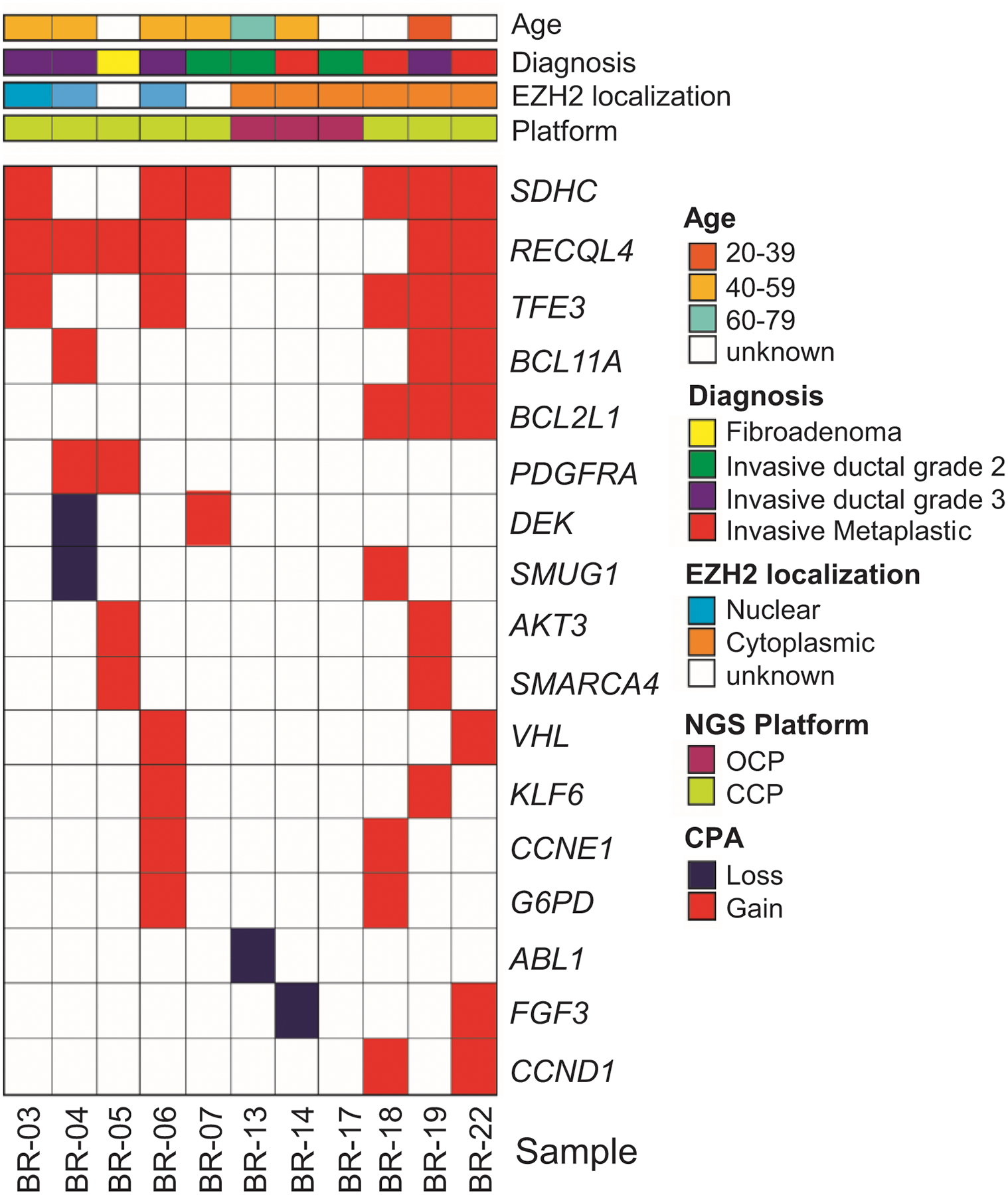
Shown are 17 high-level copy number alterations identified and color coded as indicated in the figure, using CCP and OCP NGS platforms. Ninety % of the invasive carcinomas show CNAs, largely gains, with the most frequent being RECQL4 and SDHC. Note that CNAs in PDGFRA and DEK are identified in tumors with nuclear EZH2, while CNAs in BCL2L1, AKT3, SMARCA4, CCND1, FGF3, and ABL1 are detected in tumors with cytoplasmic EZH2.
