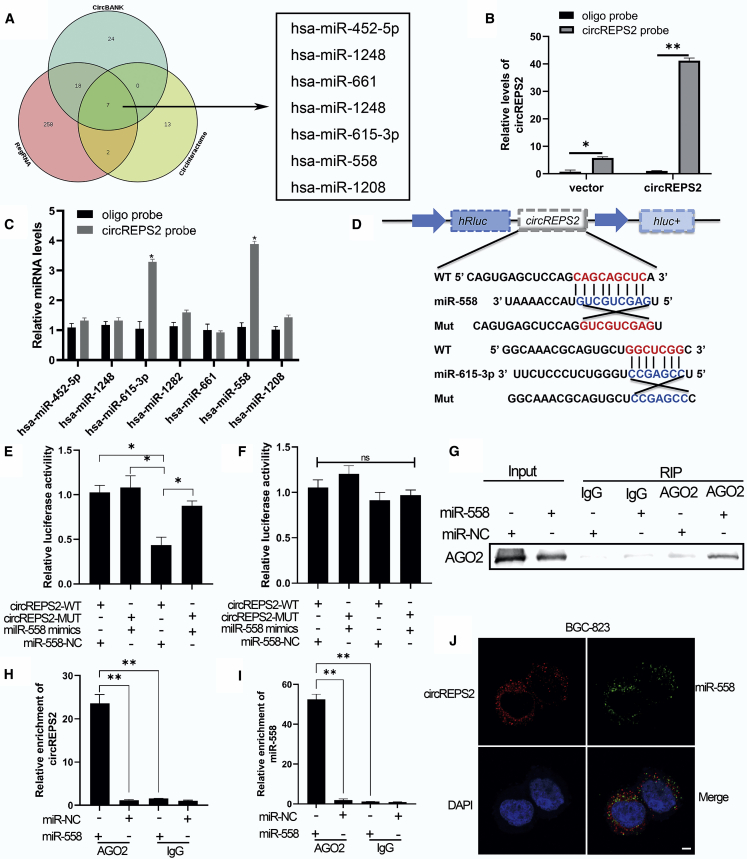
Figure 4.
circREPS2 Functioned as a miR-558 Sponge in GC Cells
(A) Schematic illustration of the overlapping target miRNAs of circREPS2 predicted by circBank, RegRNA, and CircInteractome. (B and C) RNA pull-down analysis of the enrichment of circREPS2 (B) and circREPS2-related 7 miRNA candidates in BGC-823 cells (C). (D) Schematic representation of potential binding sites of miR-558 and miR-615-3p with WT or MUT circREPS2. (E and F) The interaction between circREPS2 and miR-558 (E), miR-615-3p (F) as verified by dual-luciferase reporter assay. (G–I) Anti-AGO2 RIP analysis of the expression of AGO2 protein detected by western blotting (G), circREPS2 (H), and miR-558 (I) detected by quantitative real-time PCR in BGC-823 cells. (J) FISH analysis of the cellular colocalization of circREPS2 and miR-558 in BGC-823. Nuclei were stained with DAPI (scale bar, 20 μm). Values are shown as the mean ± standard error of the mean based on three independent experiments. ∗p < 0.05, ∗∗p < 0.01.