Figure 6.
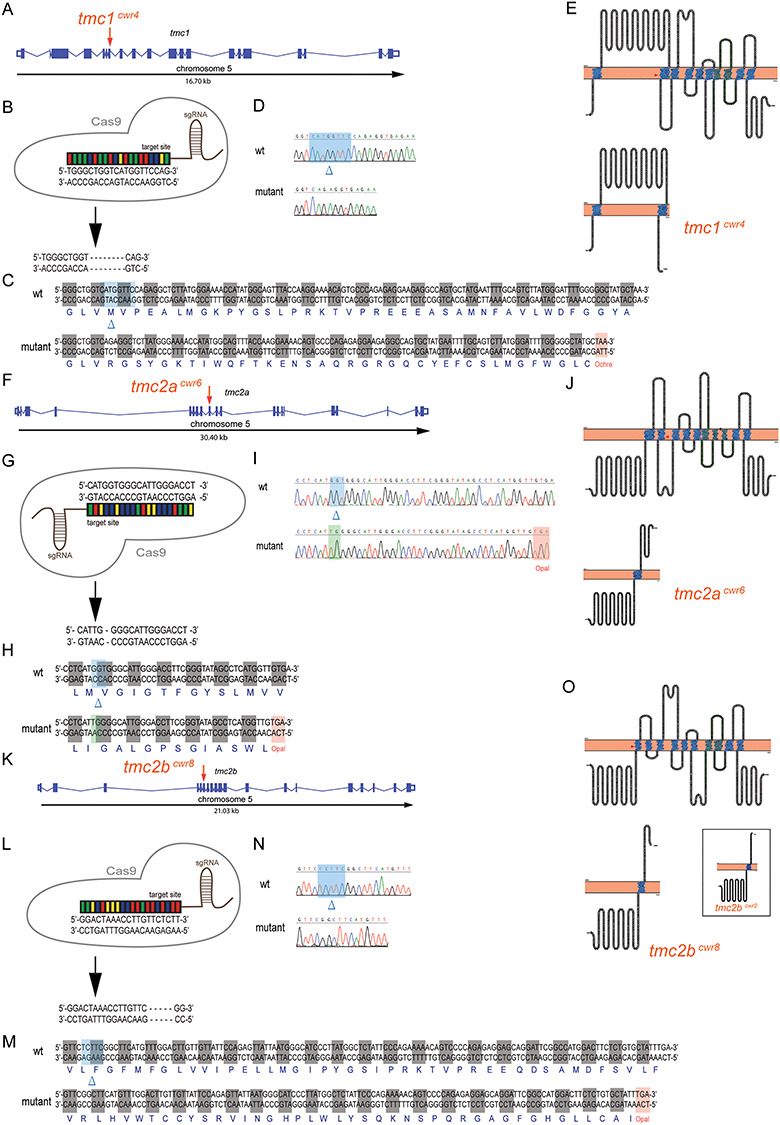
Multiplex genome editing using CRISPR to disrupt tmc1, tmc2a and tmc2b. Graphical representations of tmc1 (A), tmc2a (F) and tmc2b (K) genomic loci of zebrafish. Putative exons and splice sites are displayed. Red arrows mark the targeted exons. Segments of tmc1 exon 7 (B), tmc2a exon 9 (G) and tmc2b exon 6 (L) were subjected to genome editing. Engineered CRISPR single-guide RNAs (sgRNAs) bind target sites to enable DNA cleavage. Mutagenesis deleted eight nucleotides from tmc1 (B), deleted three and inserted two nucleotides from tmc2a (G) and deleted five nucleotides from tmc2b (L) to yield frameshift mutations. (C, H, M) Amino acid sequences of wild-type and mutant proteins. Sequencing results of mutagenized and control loci from tmc1 (D), tmc2a (I) and tmc2b (N). Blue highlights and blue deltas indicate deleted nucleotides. Red highlights denote the stop codons that were generated near the CRISPR-targeting sites. Green highlight marks the site of inserted nucleotides. (E, J, O) (Top) Topographical representations of the Tmc1 (E), Tmc2a (J) and Tmc2b (O) proteins. Arrowheads indicate points of introduced mutations. Amino acids of transmembrane domains are labeled in blue, and the TMC domains are in green. (Bottom) Schematics of predicted truncated polypeptides produced by each mutant allele are displayed. Inset, in O, displays mutant protein associated with tmc2bcwr2 produced in Chou et al 2017, emphasizing its similarity to the protein associated with tmc2bcwr8.
