FIGURE 1.
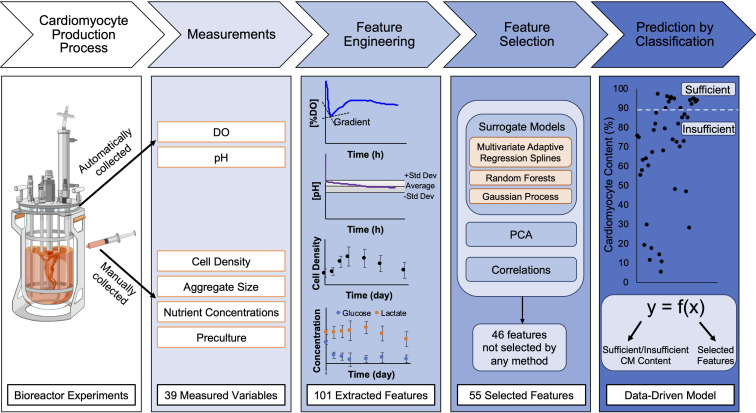
Schematic of process to generate data-driven models for prediction of final CM content from bioreactor experiments. Human pluripotent stem cells (hPSCs) were seeded in bioreactors for expansion for 48 h prior to initiation of cardiac differentiation. To initiate cardiac differentiation, a WNT amplifier, CHIR, was added for 24 h followed by 48 h of WNT inhibition using IWP2. Differentiation experiments lasted 10 days and endpoint analysis for final CM content in bioreactors was performed on differentiation day 10. During the time course of differentiation, the dissolved oxygen (DO) and pH were continually monitored. Samples were collected from the bioreactor during differentiation to analyze cell density, aggregate size, nutrient concentrations, and preculture conditions. From the 39 measured variables throughout differentiation, feature engineering was performed to extract 101 features; for example, the continuous data was separated by differentiation day to obtain averages, gradients, and second derivatives for each day. Feature selection was performed using surrogate models, principal component analysis, and correlations to determine which of the extracted features impacted the variance in the data and outcome of differentiation. After feature selection was performed, data-driven models were developed using surrogate models (MARS, RF, and GP) to predict final CM content by classification of the data in two categories (created with Biorender.com).