Abstract
To interfere with cell function, many scientists rely on methods that target DNA or RNA due to the ease with which they can be applied. Proteins are usually the final executors of function but are targeted only indirectly by these methods. Recent advances in targeted degradation of proteins based on proteolysis-targeting chimaeras (PROTACs), ubiquibodies, deGradFP (degrade Green Fluorescent Protein) and other approaches have demonstrated the potential of interfering directly at the protein level for research and therapy. Proteins can be targeted directly and very specifically by antibodies, but using antibodies inside cells has so far been considered to be challenging. However, it is possible to deliver antibodies or other proteins into the cytosol using standard laboratory equipment. Physical methods such as electroporation have been demonstrated to be efficient and validated thoroughly over time. The expression of intracellular antibodies (intrabodies) inside cells is another way to interfere with intracellular targets at the protein level. Methodological strategies to target the inside of cells with antibodies, including delivered antibodies and expressed antibodies, as well as applications in the research areas of neurobiology, viral infections and oncology, are reviewed here. Antibodies have already been used to interfere with a wide range of intracellular targets. Disease-related targets included proteins associated with neurodegenerative diseases such as Parkinson’s disease (α-synuclein), Alzheimer’s disease (amyloid-β) or Huntington’s disease (mutant huntingtin [mHtt]). The applications of intrabodies in the context of viral infections include targeting proteins associated with HIV (e.g. HIV1-TAT, Rev, Vif, gp41, gp120, gp160) and different oncoviruses such as human papillomavirus (HPV), hepatitis B virus (HBV), hepatitis C virus (HCV) and Epstein-Barr virus, and they have been used to interfere with various targets related to different processes in cancer, including oncogenic pathways, proliferation, cell cycle, apoptosis, metastasis, angiogenesis or neo-antigens (e.g. p53, human epidermal growth factor receptor-2 [HER2], signal transducer and activator of transcription 3 [STAT3], RAS-related RHO-GTPase B (RHOB), cortactin, vascular endothelial growth factor receptor 2 [VEGFR2], Ras, Bcr-Abl). Interfering at the protein level allows questions to be addressed that may remain unanswered using alternative methods. This review addresses why direct targeting of proteins allows unique insights, what is currently feasible in vitro, and how this relates to potential therapeutic applications.
Key Points
| Therapeutic antibodies are valuable drugs, which mostly act outside of cells. |
| Reaching the numerous drug targets that reside inside cells by antibodies is possible in vitro and allows unique insights compared with other methods. |
| Applying antibodies inside cells for therapeutic purposes has been explored in animal models and promises specific therapeutic benefits in neurobiology, virology and oncology. |
Why Target Proteins Directly?
Targeting proteins inside cells can serve the development of therapies in two ways: by using antibodies as drugs themselves inside cells or by using them as a tool to characterize and understand protein components of signalling pathways and thus identify targets for the development of new drugs. The hurdle for applying antibodies as drugs inside cells is often their delivery into the cell, which has so far been considered the “high-hanging fruit”, as detailed by Carter and Lazar [1]. Although DNA, RNA and proteins can all be introduced into cells, they may require different delivery methods. Delivering proteins to the cytosol is challenging, and progress has been hampered by inappropriate detection methods that substantially overestimated delivery efficiency. The importance of appropriate assays and scrutiny of evaluation criteria when interpreting the results of protein delivery experiments has since become evident [2–4].
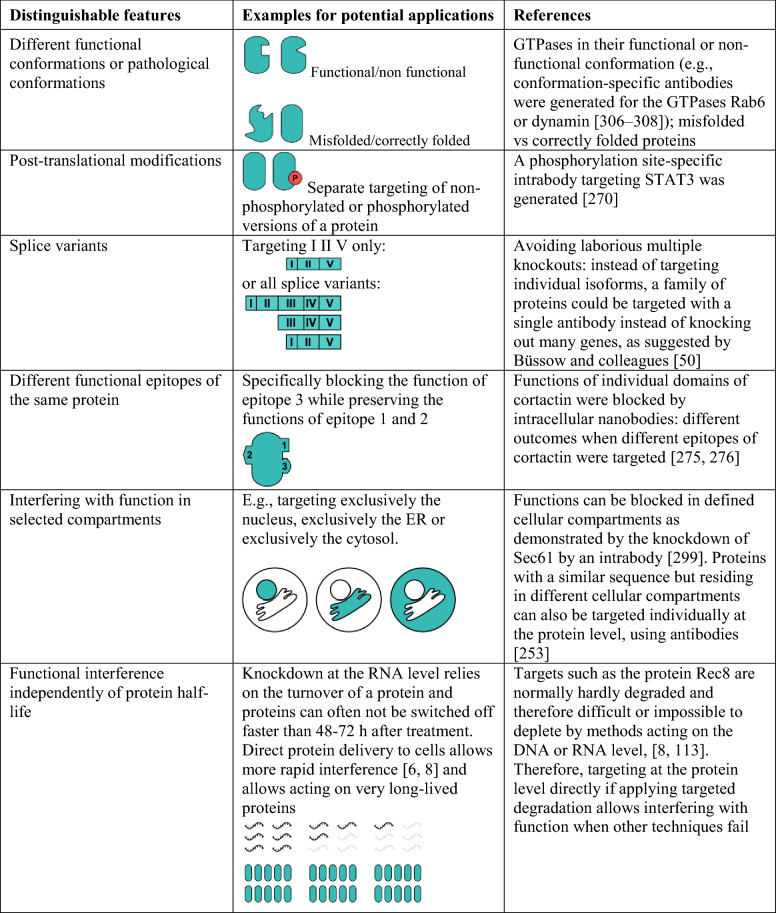
Targeting at the protein level offers the advantages of targeting protein conformations, post-translational modifications, splice variants, different functional epitopes, targeting individual cellular compartments and interference independently of protein half-life (see details in Table 1). In contrast to knocking out a gene, interference at the protein level furthermore allows tuning the amounts of a target protein instead of allowing only the presence or complete absence of the protein [5, 6]. The rapid effects achievable by protein delivery can be used to interfere in a stage-specific way and when time is a relevant factor. This is also relevant if the time required to obtain a single clone with a gene knockout is long enough to allow genetic compensation for the modification introduced [6, 7]. Delivering proteins directly circumvents stable DNA integration into the genome. The temporally more limited presence of proteins than that of permanently integrated DNA might also allow better control over therapeutic effects. Direct protein delivery is also a suitable method for inducing effects in non-dividing cells [8]. Furthermore, protein mutations could be specifically targeted and distinguished from the wild-type version, such as mutated versions of enzymes that lead to the formation of oncometabolites that may alter gene expression [9, 10]. Interestingly, not only can protein conformations be distinguished, but small antibody fragments of camelid origin (nanobodies) have even been reported to allow modification of the conformation of a target protein [11].
Table 1.
Targeting at the protein level: selected advantages
ER endoplasmic reticulum, STAT3 signal transducer and activator of transcription 3
Although small molecule drugs can target the protein level directly, not all of the proteome is druggable by small molecule drugs. Not all proteins have active sites, and protein–protein interactions are more challenging to target with small molecules [12–15]. The large surface area via which antibodies can interact with different target shapes might explain the ease with which they allow interference with protein–protein interactions.
Given its advantages, how can the protein level be targeted? Antibodies can be expressed inside cells or delivered as proteins into cells. Approaches using antibodies are reviewed here with an emphasis on long-term proven robust approaches while discussing the strengths and challenges of individual methods. Applications in the research areas of neurobiology, viral infection and immunology or oncology are reviewed.
Strategies to Use Antibodies Inside Living Cells
Antibodies can be used inside cells to neutralize targets, redirect proteins to different locations or simply analyse proteins of interest by tracking them. For instance, a membrane-tethered green fluorescent protein (GFP)-specific antibody was used to trap a GFP-fused morphogen at the cell surface, which abolished gradient formation [16]. This approach, termed morphotrap or nanotrap, demonstrates impressively how intracellular antibodies (intrabodies) can be used as tools to manipulate cellular processes by redirecting proteins of interest [16, 17]. In addition to interference, antibodies have also been used to trace and visualize proteins of interest in living cells [18, 19]. Tracing proteins not only qualitatively but also quantitatively with intrabodies allows studying protein turnover and the dynamic regulation of proteins [20]. Quantified visualization may require optimized intrabody expression [21]. Antibodies may furthermore be used to increase the turnover of a protein of interest (POI) via targeted proteolysis [6].
Sources for Binders
Molecules composed of amino acids that are able to specifically recognize another molecule are summarized under the general term ‘binder’ in the following. All binders can be used as intrabodies provided that the gene for the binder is available or the sequence is known. Depending on the compartment in which the intrabody is expressed, additional selection procedures might be required to ensure functionality of the binder in the chosen compartment [22, 23]. Methods to generate binders are briefly summarized in Table 2. For more detailed information, interested readers are referred to reviews on technologies for generating binders, such as display technologies including phage display, yeast display, ribosome display and bacterial display [24–29]. Among the group of recombinant binders, there are differences in species and in biochemical properties, which might be relevant for their use as intrabodies or other purposes. Binder species might not always be relevant if intrabodies are applied for research purposes, but therapeutic applications may require human or humanized antibodies. Certain species, such as sharks and camels, possess single-domain antibodies (sdAbs), a type of antibody that contains only a variable heavy chain (VH) but no light chain. Fragments of sdAbs of camelid origin, called nanobodies, are smaller in size than single-chain fragment variables (scFvs), a common antibody fragment of, for example, murine or human origin [30]. sdAbs may exhibit high thermal stability and the potential to refold [31, 32]. As an alternative to antibodies, designed synthetic scaffolds are potential sources for binders, such as designed ankyrin repeat proteins (DARPins), affibodies, fibronectin folds or alphaRep. Alternative scaffolds include synthetic molecules derived from ankyrin proteins (DARPins), fibronectin (e.g. monobodies), HEAT (huntingtin, elongation factor 3 [EF3], protein phosphatase 2A [PP2A], yeast kinase Tor1) protein-derived alphaRep or peptides derived from protein A (affibodies). In contrast to most natural antibodies, alternative scaffolds may not require the formation of disulfide bonds, so an additional selection procedure for functionality in the cytosol may be omitted [33–36]. The size and biochemical properties of binders may affect their suitability for intracellular use. Examples of binder formats that have already been used as intrabodies or for intracellular delivery are given in Table 2.
Table 2.
Sources of binders and their suitability for use inside cells
| Binder properties | Source for intrabody? | Source for protein delivery? | Methods to generate binders | Examples of binders | Use as an intrabody? | Use for protein delivery? |
|---|---|---|---|---|---|---|
| Polyclonal | – | + | Immunization | Full-length IgG | – | Clift et al. [8] |
| Monoclonal | – | + | Hybridoma technology | Full-length IgG | – | e.g. Clift et al. [8], Freund et al. [105], Desplacq et al. [317] |
| Recombinant | + | + | Gene isolated from a hybridoma, B cell clones or selected from libraries by display technologies | Full-length IgG | A construct of comparable size (150 kDa) containing constant domains of an IgG1 was reported as an ER intrabody [318] but not commonly used as an intrabody | e.g. Marschall et al. [3] |
| Antibody fragments | Most commonly used type of intrabodies, e.g. scFvs or nanobodies [22, 91] | |||||
| Alternative scaffolds | Examples for alternative scaffolds, e.g. DARPins, affibodies, monobodies, intracellular expression: Cetin et al. [319], Brauchle et al. [320], Vernet et al. [321] |
DARPins designed ankyrin repeat proteins, ER endoplasmic reticulum, scFvs single-chain fragment variables, – indicates no, + indicates yes
In Vivo Delivery of Binders or Binder Genes
Local in vivo electroporation has been reported for the delivery of proteins (antibodies) [37], but the in vivo delivery of proteins remains a challenge. The delivery of genes in vivo is much more advanced, with clinical trials for gene therapy in progress and some drugs already available, such as the US Food and Drug Administration (FDA)-approved gene therapy for spinal muscular atrophy (onasemnogene abeparvovec-xioi) or the in vivo gene therapy for treating an inherited eye disease (voretigene neparvovec). As a delivery vehicle for genes, adeno-associated viruses (AAVs) have been widely used, but gene therapy has also been performed with other virus types depending on the therapeutic goal, such as the FDA-approved genetically modified oncolytic virus for treating melanoma (talimogene laherparepvec) [38–43]. The therapeutic use of intrabodies is not limited to in vivo gene delivery, but intrabody genes could also be delivered ex vivo to blood cells, which could then be reinfused into the patient, as recently suggested by Png and colleagues [44]. Ex vivo gene delivery into T cells followed by reinfusion into patients has already been approved for treating B cell acute lymphoblastic leukaemia (tisagenlecleucel) [45] and large B cell lymphoma (axicabtagene ciloleucel) [46]. Ex vivo-transduced haematopoietic stem cells have been approved for treating a genetic disorder that causes immunodeficiency (severe combined immunodeficiency due to adenosine deaminase deficiency [ADA-SCID]) [47]. Gene therapy is reviewed elsewhere in more depth [43, 48–50].
Compartments Conducive to Expression or Accessible to Externally Applied Antibodies
In nature, antibodies are produced in the secretory pathway, and recombinant expression in this compartment ensures optimal conditions for correct folding. Mitochondria have also been found to be conducive to correct folding of antibodies [22, 51]. Antibodies can be directed to various subcellular compartments using appropriate sorting signals [52, 53]. Intrabodies directed to the endoplasmic reticulum (ER) can be used to knock down membrane or secreted proteins by retaining these target proteins in the ER, if an ER retention signal is fused to the antibody.
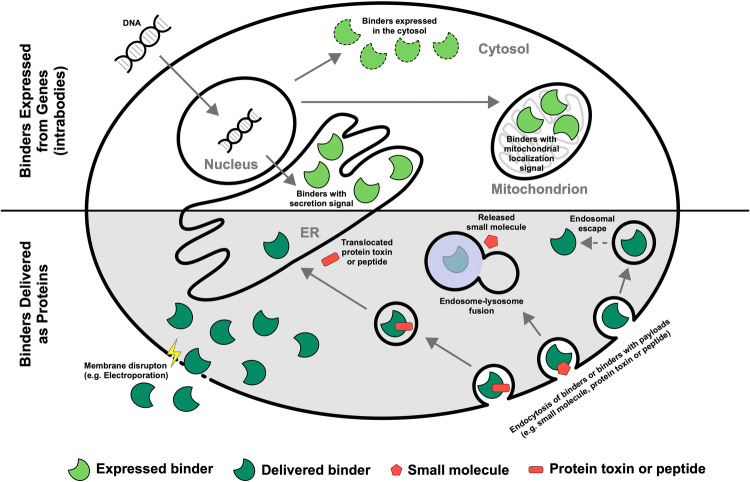
The compartments that can be targeted easily by proteins include endosomes and lysosomes. This route of delivery has, for instance, been proven successful as part of the entry mechanism of antibody–drug conjugates (ADCs) [1, 54, 55]. The various routes for macromolecules into the cell are summarized in Fig. 1.
Fig. 1.
Routes into the cell for antibodies. Antibodies can be expressed in cells or delivered as proteins to cells. Antibodies fold correctly in the endoplasmic reticulum (ER), where they are naturally expressed, as well as in mitochondria. The expression of antibodies in the cytosol may require selection for antibodies that are suitable for folding in the cytosol (dashed lines) using one of the cytosolic intrabody selection technologies. Antibodies can be delivered as proteins to the cytosol, for instance, by physical methods that are associated with membrane disruption. The critical step for antibodies that are delivered via a route that involves endosomal uptake is overcoming endosomal entrapment, which has been a major efficiency bottleneck for some of those approaches. The delivery of antibodies to the lysosome can serve to deliver small molecules to the cytosol of selected cell types, such as in antibody–drug conjugates (ADCs). Small protein toxins or peptides have been delivered to the cytosol via retrotranslocation from the ER to the cytosol
Membrane and Secreted Proteins: Knockdown with Endoplasmic Reticulum (ER) Intrabodies
ER intrabodies are antibodies that are directed into the ER by a secretory leader peptide and retained in the ER by the ER retention motif KDEL. By binding to their target protein in the ER and retaining it there, ER intrabodies are able to keep their targets away from the place where they are functional. Knockdown depends on binding only, irrespective of which epitope is bound. Target proteins can be membrane proteins, such as the type I membrane protein vascular cell adhesion molecule 1 (VCAM1), which was knocked down by an ER intrabody in vitro and in vivo [56, 57]. Targets can also be secreted proteins such as interferon-α, the secretion of which was blocked by ER intrabodies in macrophages and dendritic cells [58]. The ER intrabody approach has already been successfully applied in many research areas. Examples of the application of ER-retained intrabodies with a focus on methodological and technological aspects have been reviewed previously [22], and their therapeutic potential has been briefly discussed [59, 60].
Mitochondria-Targeted Intrabodies
A target protein has been redirected away from its natural location using an antibody that was tethered to mitochondria using a mitochondrial outer membrane targeting element [61]. Antibodies containing a mitochondrial presequence, as described by Biocca and colleagues [51], correctly form intrachain disulfide bonds, in contrast to those expressed in the cytosol. Intrabodies have been used to analyse Ca2+ dynamics at the surface of mitochondria [62]. An antibody that was targeted to mitochondria and relocalized endogenous survivin to the intermembrane space of mitochondria was suggested as a tool to study mitochondrial protein import [63]. In another report, p53 was immobilized at the mitochondrial outer membrane, resulting in reduced cell viability and morphological changes of mitochondria [64].
Antibody Delivery Routes: Endosomes, Lysosomes, ER-Associated Degradation (ERAD) and How to Use It
Delivery of proteins to endosomes and lysosomes can be easily and efficiently achieved, even if proteins often cannot escape from endosomes [65, 66]. How can antibodies be employed for therapy in these compartments? One example of the use of this cell entry route is ADCs, which consist of antibodies linked to a small molecule, such as a chemotherapeutic, kinase inhibitor or antibiotic [67–69]. While the antibody cannot cross the membrane, small molecules can, if freed from the antibody via a cleavable linker or by degradation of the antibody in the lysosome [70–72]. The proven success of ADCs in clinical practice (e.g. an ADC that was approved for the therapy of metastatic breast cancer in 2013) suggests that this route of delivery is effective.
A different delivery route is exemplified by immunotoxins, which consist of an antibody linked to a toxin, often a protein of bacterial origin [73]. Similar to ADCs, the toxin has to be released from the antibody, which can be achieved by a cleavage site [74, 75]. After release, small proteins, such as bacterial toxins can, for instance, reach the cytosol via the ER-associated degradation (ERAD) pathway [76]. Proteins are normally degraded as part of the ERAD. A low lysine content in the toxin ricin is assumed to promote the escape of some toxin molecules from degradation, which is sufficient for the toxin to be effective [77–79]. An immunotoxin for the treatment of leukaemia was approved for therapy in 2018 [55, 80]. This route of delivery is suitable for all proteins that readily refold and are effective at the dose of proteins that can escape degradation during ERAD.
The refolding of molecules translocated to the cytosol via ERAD is not necessary if the molecule is a peptide. A construct containing a peptide that can regulate immune reactions if delivered to the cytosol showed promising results in mouse models for rheumatoid arthritis [81, 82].
Compartments with No Natural Expression or Inaccessible to Externally Applied Antibodies
Cytosolic and Nuclear Intrabodies
Antibodies in the secretory compartment fold correctly, and antibodies form disulfide bonds in mitochondria. In contrast, antibodies do not form disulfide bonds in the cytosol [51], and only a few antibodies (estimated to be as low as 1% of antibodies) can be expressed as high-quality molecules in this compartment [23, 83, 84]. Approximately 0.1% of antibodies from a naive human spleen cell-derived scFv library were stable and functional [83]. Engineering a net charge was proposed as a strategy for generating antibodies that do not aggregate in the cytosol [85]. Certain frameworks may increase the chance of expression as functional antibodies in the cytosol [86–89]. Such frameworks may be selected antibody scaffolds that are functional even in the absence of disulfide bonds [90] or non-antibody scaffolds [35, 36]. The properties of nanobodies render them more suitable for expression in the cytosol [91], but various tendencies for aggregation may still occur [92, 93]. Non-antibody scaffolds may also not always be functional; only 10–20% of a fibronectin-derived alternative scaffold called FingR (Fibronectin intrabody generated with messenger RNA display) that was target-specific in vitro also co-localized with its target intracellularly [94]. Therefore, technologies such as intracellular antibody capture technology (IACT), intrabody selection after tat export (ISELATE) and “quality control” were developed to select antibodies that are suitable for cytosolic expression, which are reviewed in Marschall et al. [22, 23] and described in more detail elsewhere [83, 89, 95]. Additionally, a bacterial two-hybrid system was developed as a selection method for nanobodies as intrabodies, which has the advantage of higher transformation efficiencies and thus the chance for obtaining higher diversity than yeast two-hybrid methods [96]. To eliminate self-oligomerizing intrabodies from an intrabody library, a Fas-associated death domain (FADD) was fused to antibodies as a suicide switch against unwanted intrabodies [97]. Antibodies that are functional if expressed in the cytosol can also be directed to the nucleus if provided with the respective signal peptides. In contrast to ER intrabodies, cytosolic intrabodies need to bind a particular epitope on the target to neutralize its function [22]. Both the requirement for selecting antibodies that can fold correctly in the cytosol and the requirement for neutralizing properties make cytosolic intrabodies laborious to generate. However, cytosolic intrabodies allow promising applications, such as a recently described concept using two intrabodies against a target to sense and trigger a response to this target [98].
Protein Delivery to the Cytosol or Nucleus
A plethora of methods for the delivery of proteins have been suggested, with varying degrees of success and validation. Because one plasmid can express many proteins, low delivery efficiencies are less critical for DNA than for most proteins. A few enzymes may convert many substrate molecules, but antibodies usually have to be delivered in excess to intracellular molecules to cause an effect. Past results have demonstrated the crucial importance of choosing appropriate methods for evaluating the cytosolic delivery of proteins to avoid artefacts and the overestimation of delivery efficiency [2–4, 99].
Methods for protein delivery include approaches based on physical membrane disruption and approaches that rely on the endocytotic uptake of cargo with subsequent endosomal release [100]. For the latter class, endosomal entrapment has in the past been found to pose a major block to delivery efficiency [101, 102]. Further delivery methods include approaches that combine physical methods with chemicals, such as the application of graphene quantum dots to cells and laser irradiation [103], or use microorganisms to deliver cargoes [104]. Among the many proposed approaches to the cytosolic delivery of proteins, disruption of the membrane has proven particularly efficient for the delivery of proteins into cells [3, 105, 106]. With an efficiency of up to 90–99%, high cell viability of 80–90% [107, 108] and validation in many cell types over the last 20 years (Table 3), electroporation is a robust approach for delivering antibodies into the cytosol. Further physical methods include microinjection or microfluidic cell squeezing, which are either applicable to lower cell numbers or require special equipment [106]. Delivered antibodies remain functional in the cytosol in spite of the reducing environment. The half-life of antibodies in the cytosol is long and they bind to their targets even 3–4 days after electroporation [3, 105]. This suggests that degradation might be even slower than dilution of the antibody by cell division. Compared with DNA that has integrated into the genome, protein delivery is not permanent but transient and depends on the half-life of the delivered protein. The time-limited activity of a protein drug offers the opportunity for better control over therapy and thus higher safety.
Table 3.
Antibodies or proteins delivered by electroporation
| Delivered protein | Cell type | References |
|---|---|---|
| Asparagine synthetase antibody | HeLa, HT-5, and L5178Y DlO/R | [107] |
| p21ras antibody | B16BL6 mouse melanoma cells | [322] |
| Various antibodies | Human cells | [323] |
| MLCK antibody, constitutively active form of MLCK | Macrophages | [324] |
| Various antibodies and proteins | Pheochromocytoma, other cultured cells | [325] |
| p21ras antibody | B16BL6 mouse melanoma cells | [326] |
| Tubulin antibody | CHO cells | [327] |
| TK enzyme | TK-deficient mammalian cell line | [328] |
| Lipoxygenase antibody | Lentil protoplasts | [329] |
| Vimentin antibody, RNAse A | Fibroblasts | [330] |
| Lipoxygenase antibody | Lentil protoplasts | [331] |
| Cyclin D1 antibody | Mouse embryo and SKUT1B cells | [332] |
| TGN38-, p200- and VSV-G antibodies | NRK-6G cells | [333] |
| Tropomodulin, antibodies specific to: tropomodulin, talin, vinculin and a-actinin | Fibroblasts | [334] |
| Lucifer yellow, IgG | SW 3T3, NIH 3T3, dHL60, A7r5, BASM | [335] |
| MAP kinase antibody | MDCK cells | [336] |
| Anti-M-line protein, titin antibodies | Chicken cardiomyocytes | [337] |
| Various antibodies | Chicken cardiomyocytes | [338] |
| Wild-type STAT1, mutated STAT1, STAT1 antibody | Rat mesangial cells | [339] |
| DNase I, restriction enzymes | Jurkat cells | [340] |
| c-Src antibody | Human vascular smooth muscle cells | [341] |
| TFAR19 antibody | HeLa | [342] |
| c-Fos antibody | Spinal neuronal cells | [37] |
| STIM1 antibody | Platelets | [343] |
| Orai1 antibody | Platelets | [344] |
| EGFP | HeLa | [345] |
| Orai1 antibody | Platelets | [346] |
| HPV16 E6 oncoprotein, PCNA, RNA polymerase II largest subunit | HeLa, CaSki, H1299, MEL501 and U2OS | [105] |
| Fc-Cre, tubulin antibody, myosin antibody | SC1 REW22, HeLa | [3] |
| PCNA, DNA polymerase alpha | HeLa, US2-OS | [317] |
| Pericentrin, mTOR, IκBα, NLRP3, anti-IKKα antibodies | NIH3T3, HEK293T, human monocyte-derived macrophages | [8] |
| RBP1, TBP and TAF10 antibodies | Various mammalian or Drosophila melanogaster cell types | [108] |
| γ H2AX antibody | U2-OS | [347] |
| Various recombinant proteins | Various cell lines | [348] |
BASM bovine aortic smooth muscle, CHO Chinese hamster ovary, c-Src cellular Src, EGFP enhanced green fluorescent protein, H2AX H2A.X variant histone, HPV16 human papillomavirus 16, IKKα IκB kinase α, MAP mitogen-activated protein, MDCK Madin-Darby canine kidney, MLCK myosin light chain kinase, mTOR mammalian target of rapamycin, NLRP3 NLR family pyrin domain containing 3, NRK normal rat kidney, Orai1 ORAI calcium release-activated calcium modulator 1, PCNA proliferating cell nuclear antigen, RBP1 retinol binding protein 1, STAT signal transducer and activator of transcription, STIM1 stromal interaction molecule 1, TAF10 TATA-box binding protein associated factor 10, TBP TATA box binding protein, TFAR19 TF-1 apoptosis-related gene 19, TK thymidine kinase
Targeted Protein Degradation
Targeted proteolysis as an alternative to RNA interference (RNAi) and CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9-based knockdown allows rapid interference with protein levels, which is independent of regulation on the RNA and DNA level. While tools for performing knockdown via RNAi or CRISPR/Cas9 have so far been accessible more easily, the field of targeted protein degradation has recently undergone rapid progress. Targeted proteolysis can be mediated both by intrabodies (antibodies expressed intracellularly) and delivered antibodies (protein delivery). Two types of targeted proteolysis are distinguished here: approaches using degrons and approaches using high-affinity binders (Table 4).
Table 4.
Comparison of strategies for targeted degradation
| Type | Module to target for degradation | Requirements and features |
|---|---|---|
| POI fusion | Degron | Modification of the POI and genetic modification of the cell/organism required |
| High-affinity binder based (unmodified POI) | TRIM21 | Binder with a constant (Fc) domain has to be used, protein delivery required |
| Bispecific small molecule (PROTAC) | Availability of a small molecule binder for the POI required | |
| Binder fused to a degron | Either expression of the binder from a gene or protein delivery can be used; no Fc domain is required, and therefore more choice is available for the binder format/type (e.g. IgG, scFv, nanobody, alternative scaffolds) |
POI protein of interest, PROTAC proteolysis-targeting chimaera, scFv single-chain fragment variable, TRIM21 tripartite motif containing-21
Targeted Proteolysis by Modifying the Protein of Interest (POI)
Degrons enable proteolysis by fusing a short sequence to the POI, which means that the POI needs to be modified. Proteolysis approaches based on high-affinity binders may use macromolecules or small molecules as binders and can act on endogenous, unmodified POIs. The proteolysis of degron-tagged proteins can be controlled by adding a small molecule that either induces degradation upon addition or induces degradation upon withdrawal. The auxin-inducible degron (AiD), a proteolysis approach based on a plant hormone, allows degradation of the degron-tagged POI upon the addition of a small molecule. Upon fusing a POI with a SMASh (small molecule-assisted shutoff)-tag, the SMASh-tagged POI can also be degraded upon addition of a small molecule, but because of its special property of promoting the proteolysis of newly synthesized proteins upon addition of the chemical, this degron allows research on the protein half-life and degradation of the POI. In contrast, for FKBP12 (12 kDa FK506-binding protein)- or UnaG-tag based degrons the degron-tagged POI is stabilized upon addition of the small molecule and degraded as soon as the small molecule is absent [5, 6, 109–111].
Targeted Proteolysis Via High-Affinity Binders: Keeping the POI Unmodified
High-affinity binders allow the targeted proteolysis of unmodified POIs. Proteolysis-targeting chimaeras (PROTACs), which are bispecific binders, have recently gained much attention, with the first one reaching clinical studies [112, 113]. The concept of bispecificity is well-known for intercellular applications, such as bispecific antibodies that link tumour cells and immune cells to promote the killing of tumour cells [114]. The same concept can also be used for intracellular applications. By recruiting the cellular degradation machinery to a POI, degradation of the chosen protein can be achieved. The first PROTACs, which were based on peptides, suffered from a lack of cell permeability. More recent generations of PROTACs were constructed from small molecules and have shown improved cell permeability [113, 115]. Because PROTACs keep recruiting POIs, a single PROTAC molecule can induce the proteolysis of many POI molecules [116]. PROTACs have already been used successfully in vivo, and an oral PROTAC drug has been approved for a phase I clinical trial as a drug for the treatment of prostate cancer. In spite of the great success of PROTACs, future work is still needed; for instance, Guo and colleagues [117] state that there is still a “challenge for designing on-target PROTACs” [118]. Although small-molecule PROTACs are cell permeable, in contrast to antibodies, the high specificity and almost unlimited diversity of proteins that can be targeted by antibodies make them an attractive tool for targeted degradation. More than 30 proteins have already been targeted by PROTACs [115], but hundreds of antigens can already be targeted via antibodies derived from phage display with HAL libraries alone [24], not including the enormous number of antibodies from other sources allowing access to the antibody gene. The technology for generating antibodies specific for a new POI is a well-established and robust procedure [24]. The high number of proteins that can be targeted with antibodies can therefore justify the trade-off between diversity of targeted antigens and deliverability that is more favourable in small-molecule PROTACs.
Antibodies can, for instance, be delivered into cells via electroporation or microinjection. Delivering full-length IgGs to cells has been described as a means to achieve the degradation of a POI dependent on the tripartite motif protein TRIM21 (tripartite motif containing-21)/Ro52 [8]. TRIM21/Ro52 first attracted attention due to its particular antigenicity and as a target for autoantibodies in autoimmune diseases [119]. TRIM21/Ro52 is an E3 ubiquitin ligase that has been proposed to act as a mechanism of intracellular antiviral defence by binding to the Fc part of antibodies attached to viruses that enter the cytosol and subsequently induce degradation of antibodies and viruses via the proteasome [120]. Targeted degradation via TRIM21 can therefore be achieved using full-length antibodies or by using proteins fused to the Fc part of an antibody. While all cells can express TRIM21, it is expressed at different levels in different cell types [8]. The efficiency of targeted degradation depends on the expression level of TRIM21 as well as the amount of antibody and the amount of target present. TRIM21-mediated targeted proteolysis was reported to be rapid, occurring within minutes [8]. Although endogenous TRIM21 levels may be sufficient in some cells, TRIM21 has to be overexpressed or delivered as a protein to mediate proteolysis sufficiently in other cells [8]. Using antibody- and TRIM21-mediated proteolysis, a method termed ‘Trim away’, allowed depletion of the long-lived protein Rec8, which has a role in sister chromatid cohesion and does not turn over for long time periods in mice and possibly for years in humans. If there is a lack of turnover of the POI, depletion via RNAi would be ineffective. Gene knockout is furthermore not possible if a gene is essential for viability [8, 121, 122]. Furthermore, by using antibodies that specifically recognize a pathogenic version of the protein huntingtin, Trim away allowed selective depletion of only the pathogenic protein variant that was co-expressed with the normal huntingtin protein [8]. This demonstrates the possibility of selectively depleting protein variants, which could also be employed for selectively depleting proteins with post-translational modifications.
Targeted degradation was also mediated by expressing antibodies in cells instead of delivering them as proteins. The binder is genetically engineered for targeted proteolysis, leaving the POI unmodified [6]. Selecting antibodies that are functional if expressed in the cytosol is often more elaborate than normal antibody generation procedures, but the use of an anti-GFP antibody allowed a single antibody to target many antigens for degradation if GFP-tagged [123]. This approach is especially attractive for model organisms with GFP-tagged proteins, such as those often used in developmental biology. In zebrafish and flies, this approach has already been successfully applied [124–126]. Various tags and motifs can be employed to target antibodies and their antigens for degradation, such as providing the binder with an AiD, a proline, aspartate or glutamate, serine and threonine (PEST) motif, the Von Hippel Lindau protein, Speckle-type POZ protein (SPOP), an F-box domain or the catalytic domain of a ubiquitin ligase [6, 124, 127–132].
A nanobody fused to an AiD allowed inactivation of the anaphase-promoting complex/cyclosome (APC/C) in vitro and in zebrafish. Genetic engineering was required to allow expression of the nanobody–degron fusion, and this approach also required expression of the plant F-box protein, protein transport inhibitor response 1 (TIR1). The advantage of this approach was its reversibility as well as rapid and effective temporal control over degradation via auxin supply. Marked degradation occurred within 16–28 min, and protein levels had recovered sufficiently to rescue the knockdown phenotype 8 h after the removal of auxin [131]. An intrabody fused to an F-box domain was used to specifically degrade the active form of the GTPase RHOB [132]. ‘Ubiquibodies’ are ubiquitin ligases in which the natural substrate binding domain has been replaced by an antibody, which allows the targeting of POIs for degradation [127]. Integrating PEST motifs into the sequence of a binder has also been used for the purpose of targeted degradation [130, 133, 134], so, overall, there is a wide range of strategies and tools available to target proteins for degradation once an antibody can access a POI.
Targeted Proteolysis Via Lysosomes or the ER
The approaches mentioned so far employed the cytosolic degradation machinery, but approaches to perform targeted degradation in other compartments have also been described. Lysosome-targeting chimaeras (LYTACs) are based on the strategy of employing the cation-independent mannose-6-phosphate receptor (CI-M6PR) to target POIs for lysosomal degradation. LYTACs consist of an antibody that is fused to a ligand for this receptor and allows targeting membrane proteins and secreted proteins for lysosomal degradation [135]. Further strategies for lysosomal targeting include approaches based on receptor crosslinking and targeting to the lysosome by means of peptides [65, 66]. Degradins allow targeting of proteins in the ER for degradation. Degradins consist of a moiety that is specific for the POI and the protein SEL1L, which is involved in identifying misfolded proteins in the ER to subject them to the ERAD pathway. Degradins were thus proposed as a strategy to retrotranslocate POIs from the ER to the cytosol for proteasomal degradation [136].
Applications in Neurobiology
Many neurodegenerative diseases are associated with protein misfolding and aggregation [137]. There is controversy about which forms of proteins are toxic in neurodegenerative disorders. While some studies attribute toxicity to the aggregated form, other studies suggest aggregation to be a beneficial process that protects the cell from intermediate or misfolded toxic proteins [138, 139]. Neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) are also discussed as potential prion-like diseases for which misfolded proteins are known to propagate and convert other proteins into pathological forms [140, 141]. Because protein aggregation occurs downstream of the gene level, studying neurodegenerative diseases requires methods that allow direct targeting of proteins. To date, there is no cure for neurodegenerative diseases, and all available treatments only manage symptoms or halt disease progression. Drug repurposing, which is the evaluation of existing drugs normally used to treat other diseases such as cancer, asthma, infections and others for their therapeutic potential in treating neurodegenerative diseases, is an active area of research with the advantage of already pre-existing knowledge about the pharmacokinetic and pharmacodynamic profiles of drugs [142]. Beyond repurposing existing drugs, further research on molecular mechanisms and potential new therapies for neurodegenerative diseases is desirable. With their suitability for targeting different conformations and aberrant versions of protein structures, antibodies are particularly interesting as research tools or as potential new therapies for protein misfolding diseases (Table 5).
Table 5.
Intracellular antibodies applied in research and as potential therapeutic strategies
| Research area | Disease/disease process | References |
|---|---|---|
| Neurobiology | Alzheimer’s disease | [152–157, 161, 163, 165, 349] |
| Parkinson’s disease | [130, 136, 145–148] | |
| Huntington’s disease | [133, 168, 170–175, 310, 350, 351] | |
| Amyotrophic lateral sclerosis | [182, 184] | |
| OPMD | [185] | |
| Viral infections | HIV | [213, 215, 217, 218, 221, 222, 224–227, 229] |
| EBV (HHV-4) | [235, 236] | |
| HHV-8 | [238, 241] | |
| HBV | [259–261, 352]÷ | |
| HCV | [265–267, 270, 353] | |
| HPV | [246–251] | |
| Rotavirus | [199] | |
| Influenza A | [202, 203] | |
| Ebola | [209, 354] | |
| Marburg virus | [209] | |
| Bluetongue virus | [355] | |
| Hantavirus | [356] | |
| Vesicular stomatitis virus | [357] | |
| Rabies | [358] | |
| Porcine viruses | [359, 360] | |
| Maedi visna virus | [361] | |
| Flavivirus | [362] | |
| Cucumber mosaic virus | [194, 196] | |
| Cancer research | Cellular level: oncogenic pathways, proliferation, cell cycle, apoptosis | [64, 274, 276, 278, 363, 356, 364–369] |
| Tissue level: adhesion, metastasis, angiogenesis | [132, 279–288, 370–377] | |
| Neo-antigens | [291, 295, 296] |
EBV Epstein-Barr virus, HBV hepatitis B virus, HCV hepatitis C virus, HHV human herpesvirus, HPV human papillomavirus, OPMD oculopharyngeal muscular dystrophy
Parkinson’s Disease
PD, the second most common neurodegenerative disease, is characterized by the loss of dopaminergic neurons, and is best known for causing motor symptoms. PD is associated with misfolded forms of the protein α-synuclein, which therefore has been considered of interest as a potential drug target [143]. Several α-synuclein intrabodies and engineered versions thereof have been described to reduce aggregation and toxicity in vitro [144–148]. In vivo, gene therapy using AAV-based vectors has been performed to deliver α-synuclein intrabodies into rats. Because antibodies can be employed to target selected regions of a protein, intrabodies can be used to map structure function relationships in detail. This advantage was used by employing two different intrabodies, targeting the non-amyloid or the C-terminal part of α-synuclein. The two different intrabodies were further engineered with a PEST motif for proteasomal degradation. One intrabody showed marked maintenance of striatal dopaminergic tone compared with controls. Furthermore, modest behavioural rescue was observed, although pronounced variability occurred among individual animals. While authors mention the importance of evaluating the model system as well as the timeline of therapeutic intervention, this study demonstrates the feasibility and potential of intrabody gene therapy in vivo [130].
Alzheimer’s Disease
Gene therapy to express antibodies in animal models has also been applied multiple times as a potential new treatment for AD and was reviewed in depth by Cardinale et al. [149] and Meli et al. [150]. AD is the most common neurodegenerative disease causing dementia. It is associated with the aggregation of amyloid-β (Aβ) peptide, which therefore raised interest as a potential drug target [151]. The Aβ peptide is formed via proteolysis of the Aβ precursor protein (APP), and intrabody binding to an epitope adjacent to the cleavage site reduces Aβ formation by shielding the cleavage site. Another even more efficient intrabody almost completely prevented Aβ formation by inducing disposal of APP from the ER [152]. As a complementary approach, an intrabody was used to suppress the function of a protein involved in APP proteolysis [153]. Instead of targeting cleavage, the different products of cleavage can also selectively be distinguished by intrabodies. Misfolding leads to the formation of pathological aggregates, as has been demonstrated using conformation-specific intrabodies. This opens up promising new therapeutic avenues, since conformation-specific intrabodies could target only those Aβ conformers that undergo pathological oligomerization without interfering with the ‘healthy’ processing of APP [154]. The intrabody could thus act as a filter that allows only ‘healthy’ conformers to pass. Gene therapy via the AAV-mediated delivery of an anti-Aβ42 intrabody in an AD mouse model allowed partial clearance of Aβ42 deposits [155], and in a follow-up study, myelin integrity was restored in mice treated with intrabody gene therapy [156]. An intrabody was furthermore used for imaging purposes in live neurons to study mechanisms of memory impairment in AD [157], and intrabodies have been found to be generally valuable tools for imaging purposes in neurons [158].
AD is also associated with abnormal tau phosphorylation as well as aggregation and is the most common tauopathy [159, 160]. Melchionna and Cattaneo [161] reported the degradation of tau by intrabody-mediated tumour necrosis factor (TNF)-α inducible targeted proteolysis. However, this approach might be limited as a therapeutic approach due to the adverse effects of the cytokine TNF-α and is more likely to serve as a research tool [162], as reviewed by Messer and Butler [160]. Targeted degradation of tau via expressing an AAV-delivered intrabody resulted in a statistically significant decrease in tau-associated pathology in a human-tau transgenic mouse model as reported by Gallardo and colleagues [163]. Among several splice variants of tau, the 2 N isoform has been suggested to preferentially interact with proteins that are associated with neurodegenerative diseases [164]. The effect on a selected isoform could be imagined to be more effective if targeted by an antibody specific for only an individual splice variant compared with a drug that recognizes all isoforms because binding does not compete with the other isoforms. A combination of focused ultrasound and administration of an antibody specific for the 2 N isoform of tau was applied to deliver the antibody to the brains of mice. This treatment was found to reduce tau hyperphosphorylation and anxiety-like behaviour in a transgenic mouse model that overexpresses the 2 N isoform of tau [165].
Huntington’s Disease
Huntington’s disease (HD) is a hereditary disorder caused by a mutant version of the huntingtin gene (mHtt), which is associated with motor symptoms and cognitive decline as a consequence of the death of brain cells in the striatum [166]. If gene therapy is used to target mHtt as a treatment for HD, the pathologic protein could still remain in the body and cause pathologic effects due to its ability to act as a seed and convert other proteins into pathologic molecules [167]. For this reason, even in the absence of the mutated gene, the pathological protein would have to be removed, which could be performed with intrabodies. Multiple studies have already focused on intrabodies as a promising treatment option for HD, which are reviewed in detail by Denis et al. [168, 169]. In vitro, intrabody expression resulted in decreased mHtt aggregates or increased mHtt turnover and increased cell survival. Therapeutic effects were enhanced when drug administration was combined with intrabody expression in vivo in a fly model [170]. In transgenic mice or mice that received intrabody gene therapy, decreased motor and cognitive impairments and increased body weight and lifespan were observed [169, 171–175].
Further Applications in Neurobiology
Amyotrophic lateral sclerosis (AML) is a neurodegenerative disease characterized by motor deficits, which can also be associated with cognitive and behavioural changes. In most cases the mechanisms causing the disease are unknown, called sporadic ALS, but in a subset of patients the disease is familial involving mutations in the genes C9orf72, TARDBP (TDP43), FUS and superoxide dismutase (SOD1) among others [176–180]. One hypothesis is that the degeneration of motor neurons in AML caused by mutated versions of SOD1, might be associated with the tendency of mutant SOD1 versions to misfold and aggregate [176]. Ghadge and colleagues [181] generated two anti-SOD1 antibodies, which prolonged survival in a mouse model after delivery by gene therapy. Gene therapy was also performed to express antibodies specific for misfolded SOD1 in mice, resulting in delayed onset of ALS pathogenesis and an extended lifespan [182]. TDP43 is an RNA/DNA binding protein involved in the regulation of gene expression and RNA processing. TDP43 aggregates not only in ALS but in other neurodegenerative diseases such as frontotemporal lobar degeneration (FTLD), therefore being a promising target for multiple diseases [183]. An antibody specific to a selected domain of TDP43, RNA recognition motif 1 (RRM1), was generated by Pozzi and colleagues [184] to obtain two therapeutic effects at the same time. The TDP43-specific antibody should interfere with protein aggregation and additionally combat hyperactive inflammatory responses by blocking protein–protein interactions of the RRM1 domain. Gene therapy of mice with this antibody had a protective effect and could be a novel therapeutic avenue for AML and for TDP43 tauopathies in general [184].
Oculopharyngeal muscular dystrophy (OPMD), a protein aggregation disease that is characterized by the weakening of specific muscle groups involved in holding the eyelids and swallowing and of proximal limb muscles, was nearly completely rescued in vivo by an intrabody in a fly model [185].
Apoptosis might be responsible for cell death in several neurodegenerative diseases. Inhibition of the proapoptotic protein Bax was proposed as a means to confer resistance of cells against apoptosis [186].
The optimization of intrabodies to treat neurodegenerative disorders was reviewed in depth in a recent publication by Messer and Butler [160].
Applications in Viral Infections
Intrabodies used in the research area of viral infections belong to the class of cytosolic intrabodies or the class of ER intrabodies, if the proteins of interest are in the secretory pathway [58, 187–193]. The intracellular use of antibodies has been considered a valuable option by researchers for targeting many viruses (Table 5). The ease with which plants can be genetically modified allows conferring an antibody-based ‘immune response’ on plants. Intrabodies were expressed in tobacco and tomato plants as a means of protecting them from infection with the cucumber mosaic virus, which has economic relevance due to its wide host range and worldwide occurrence [194–196]. Antibodies from camelid origin have also been reported to be expressed in plants [197, 198].
Understanding the mechanistic details of infections through intrabody-mediated interference may help with identifying new strategies for pharmaceutical intervention. An advantage of applying intrabodies in viral infection research is the possibility of disrupting viral protein function without genetic manipulation of the virus itself. For instance, an intrabody was used to study rotavirus [199], a non-enveloped RNA virus causing gastroenteritis [200]. Intrabodies also allowed the mechanistic details of influenza A, an enveloped RNA virus, to be studied [201–203]. Because genetic engineering often impairs or abrogates viral function for small RNA viruses such as influenza A [202, 204–206] and inhibition by chemicals is not always possible, intrabodies allow questions to be answered that cannot be addressed by these methods. Beyond analysing virus biology, intrabodies were also used in a more therapy-oriented approach. Infection with Ebola and Marburg virus, which are both enveloped RNA filoviruses [207], poses a significant threat due to their high lethality rates of up to 90% in some cases of outbreaks [208]. As a strategy to allow only empty and non-infectious particles to form by inhibiting packaging, viral proteins from Ebola and Marburg virus were cross-linked inside the cell via a dimeric intrabody [209].
Chronic Viral Infections and Oncoviruses
Human Immunodeficiency Virus
Human immunodeficiency virus (HIV) is an enveloped (+)-strand RNA retrovirus that can eventually cause acquired immunodeficiency syndrome (AIDS), causing distinct depletion of CD4+ T cells and thus death by progressive failure of the immune system if not treated. A combination of antiretroviral drugs is used to manage the disease as a chronic condition to prevent the development of AIDS in patients [210–212]. Similar to the approach of combination therapies used in HIV treatment, intrabodies were used to target different points in the infectious cycle. Both viral and host proteins have been targeted by intrabodies to attack HIV, including the matrix protein p17 [213], the viral surface protein gp120, which is responsible for binding to the cellular receptor [214, 215], gp41, which is involved in fusion of virus and host membranes [216, 217], the virion infectivity factor Vif, which counteracts antiviral mechanisms of the host [218, 219], Rev, which is responsible for the export of viral RNA from the nucleus [220, 221], the viral accessory protein Vpr, which disturbs cellular pathways [222, 223], the viral protein TAT, which allows viral gene expression [215, 224–226], the cellular protein LEDGF (lens epithelium-derived growth factor) [227], and cellular receptors involved in viral entry of HIV into cells, such as the host proteins CCR5 (C–C chemokine receptor type 5) [189, 191, 193] and C-X-C chemokine receptor type 4 (CXCR4) [190, 192, 228]. An intrabody fused to a fluorescent protein was further used to allow dynamic tracing of HIV in cells [229]. To interrupt viral replication by allowing only non-infective virions to form, intrabodies were expressed that were targeted for degradation and specific for gp160 [230]. Because of its high mutation rate, HIV continues to change in patients, which can result in drug resistance [231]. A way to circumvent this problem could be targeting host proteins that are relevant in the viral life cycle, but do not have high underlying mutation rates like the viral proteins. Protein–protein interaction between a viral and a host protein was blocked by an intrabody as a means to interfere with viral replication and proposed as a potential intracellular immunization for T cells in HIV-positive patients [227]. By expressing an ER intrabody that retained the host cell surface receptor CCR5 in the ER of CD4+ T cells, these cells were protected from viral entry both in vitro and in a mouse model [193]. Because this approach is also based on targeting a host protein, the ER intrabody is not at risk of losing the ability to bind its target, as might occur with escape mutants if a viral protein is targeted. To address problems with drug resistance and residual infected reservoir populations under antiretroviral therapy, alternative approaches such as gene therapy could be further explored to improve therapeutic efficacy. Viral gene expression is stimulated by the HIV-1 Tat protein [232]. In a preclinical study with rhesus macaques, CD4+ T cells were isolated from macaques for ex vivo transduction with an HIV-1-Tat-specific intrabody to promote the survival of CD4+ T cells. Modified CD4+ T cells were re-infused into macaques, and the animals were subsequently challenged with the virus. CD4+ T cells expressing the Tat-specific intrabody survived longer than cells expressing a control intrabody after challenge, and a reduced viral load was observed in one of two animals [226].
Epstein–Barr Virus
Epstein–Barr virus (EBV), also called human herpesvirus 4 (HHV-4), is a DNA virus that may cause cell transformation and has a global prevalence of more than 90%. Although EBV infection is in most cases lifelong dormant, EBV was the first virus identified as an oncovirus and is associated with cancers such as lymphomas and epithelial cancer and is a risk factor after transplantation for developing post-transplant lymphoproliferative disease (PTLD) [233, 234]. Latent membrane protein 1 (LMP1) of EBV is essential for B cell transformation, and its knockdown was achieved by expressing an ER intrabody. By reducing LMP1 levels via an ER intrabody, cells were rendered more sensitive to chemotherapeutic-induced cell death [235]. The same target, LMP1, was also attacked at the cytosolic part of the LMP1 transmembrane protein by a cytosolic intrabody. To induce a functional effect with a cytosolic intrabody, an antibody was generated against a selected epitope of LMP1, which serves as a docking site for downstream signalling molecules [236]. This demonstrates how the same protein can be targeted in different ways by intrabodies and how the experimental approach differs accordingly. As a therapeutic strategy, EBV-infected tumour cells could be sensitized to chemotherapeutic drugs by ER intrabodies or by cytosolic intrabodies as gene medicines [235, 236].
Human Herpesvirus 8
Kaposi sarcoma-associated herpesvirus, also called human herpesvirus 8 (HHV-8), has a global prevalence of less than 10% but a regional prevalence of up to 70%. HHV-8 is an enveloped DNA virus that is usually asymptomatic upon primary infection but can cause Kaposi sarcoma in immunodeficient individuals [234]. Latency-associated nuclear antigen (LANA1) helps to maintain the viral DNA of HHV-8 in daughter cells [237] and was therefore chosen as a promising drug target for intracellularly expressed antibodies [238]. A cancer-promoting feature of HHV-8 is its expression of proteins that are very similar to host proteins and it can therefore interfere with regulation of the host’s cell cycle, apoptosis and cytokine signalling. Viral interleukin-6 (vIL6) mimics the activities of human interleukin-6 (hIL6), but stimulation by hIL6 depends on hIL6 receptor (hIL6R), while vIL6 can stimulate cells even independently of hIL6R [239, 240]. By the expression of an ER intrabody that retained vIL6 in the ER, the secretion of vIL6 was prevented, and vIL6-mediated signalling was blocked [241].
Human Papillomavirus
Human papillomavirus (HPV) has a prevalence of 70 million cases in the USA alone. Each year, an estimated 500,000 cases of cervical cancer occur, which are caused by high-risk HPV strains. The first prophylactic vaccine against high-risk HPV strains was approved in 2006, but only part of the global female population is vaccinated [242]. HPV belongs to the non-enveloped DNA viruses, and the viral proteins E6 and E7 have oncogenic activity or contribute to malignant progression [243]. In high-risk HPV strains, E7 has transforming activities, and the tumour suppressor p53 can be inactivated by E6, preventing p53 from inducing apoptosis, which makes both proteins interesting as drug targets [244, 245]. There have been reports of using intracellularly expressed antibodies to influence oncogenic effects of the viral proteins E6 as well as E7 [246–250]. Interestingly, the epitope recognized by an antibody directed against E6 was found to be in a region that could hinder recognition of the E6 nuclear export signal. As hypothesized by the authors, this could be a potential mechanism by which export of E6-bound p53 to the cytosol might be blocked, and, as a consequence, proteasomal degradation of the tumour suppressor p53 might be prevented [249]. The strategy to influence the subcellular localization of a protein for modulating its function, or as hypothesized here to influence turnover as a consequence of trapping in the nucleus, allowed studying cells in an entirely new way. The prevention of p53 degradation was also achieved in HPV-infected cells by an intracellularly expressed nanobody. In contrast to the aforementioned example, the intracellular nanobody did not bind E6 but was specific for the DNA binding domain of p53. Although the tumour suppressor p53 was stabilized against degradation by the nanobody, the function of p53 was inhibited upon expression of the nanobody, resulting in increased cell proliferation [251]. This impressively demonstrates how intrabodies can be used to study different functional details of the exact same process, in this case by targeting a different binding site. The therapeutic potential of intrabodies in HPV treatment was further explored by Amici and colleagues [250]. An E6-specific intrabody was isolated using IACT, a method for selecting antibodies that fold correctly in the cytoplasm. The E6-specific intrabody was expressed and directed to the nucleus in cells. As a result, p53 accumulated in the nucleus of SiHa cells and was able to partially inhibit its degradation. Via a retroviral vector, the E6 intrabody was expressed in preclinical mouse models of HPV and resulted in a marked delay of tumour onset. While all mice had developed tumours by 20 days after injecting HPV16-positive tumour cells, 60% and 40% of mice injected with TC-1 and C3 intrabody-expressing tumour cells, respectively, remained tumour free for more than 4 months [250].
Hepatitis B Virus
Hepatitis B virus (HBV) is an enveloped DNA virus with a virion diameter of approximately 45 nm and a 36 nm diameter core [252]. HBV can be prevented by prophylactic vaccines, but vaccination is not sufficiently effective to induce seroprotection for every person. Approximately 5% of the population does not respond sufficiently to vaccination [253] and HIV patients in particular have lower response rates [254, 255]. The regional prevalence of HBV infection varies [234, 256]; with a prevalence of 10–25% in some developing countries, HBV is still a concern [257]. HBV infection can result in various liver disorders or hepatocellular carcinoma, which still causes 500,000–700,000 deaths per year [253, 258, 259]. Expression of a nanobody as an intrabody in the cytosol or nucleus was used to attack the HBV core protein (HBcAg) [260]. The secreted viral proteins hepatitis B surface antigen (HBsAg) and hepatitis B e-antigen (HBeAg) have been retained in the ER by ER intrabodies. HBeAg secretion was downregulated by the ER intrabody, but HBcAg was not affected, although its sequence is similar, and cross-reactivity of the anti-HBeAg antibody to HBcAg was shown [261]. This demonstrates how proteins with high sequence similarity but residing in different compartments can be targeted separately from each other by employing the compartment-specific action of intrabodies. An ER intrabody specific for HBsAg was shown to inhibit secretion of HBV virions in vivo in a mouse model, demonstrating the therapeutic potential of this approach [259].
Hepatitis C Virus
Hepatitis C virus (HCV) causes approximately 400,000 deaths per year as a consequence of chronic HCV-induced liver disease and liver carcinoma, in spite of advances in HCV treatment [262, 263]. HCV is an enveloped (+)-strand RNA virus that produces viral components as a polyprotein that needs to be cleaved into its individual functional protein units [264]. HCV core protein, which forms the viral capsid [264], was the target of intracellularly expressed antibodies [265–267]. Despite the availability of direct-acting antivirals and small-molecule inhibitors for HCV treatment, the high mutation rate of HCV can rapidly lead to the emergence of drug resistance in patients during treatment [268, 269]. The inhibitory effect observed upon expression of an NS3-specific intrabody was maintained even in the presence of point mutations that confer resistance to small-molecule drugs. As hypothesized by the authors, resistance to antibody-based drugs might occur more slowly than resistance to small-molecule-based drugs because antibodies contact their targets over a comparably large surface area via multiple residues [270].
Targeting Host Processes in Viral Infections
Viral infections are counteracted by the host protein interferon (IFN)-α, but due to the high homology of the 14 different isoforms of IFN-α in mice, generating knockout mice on a genetic level would require immense effort. A single intrabody that recognizes all isoforms could provide a solution. Following this hypothesis, an ER intrabody specific for IFN-α isoforms was expressed in different cell lines in vitro and allowed blocking of IFN-α secretion as well as increased virus proliferation as a consequence of ER intrabody-mediated IFN-α knockdown [58]. Knocking out many genes with similar functions to generate a mouse model may no longer be required if all isoforms of a protein can be targeted with a single intrabody, which means only a single gene has to be integrated into the genome to generate a transgenic intrabody mouse. Because intrabody expression is dominant, a marked phenotype may already be observed in an earlier stage during the generation of a mouse model: heterozygous intrabody mice might already display sufficient knockdown of the target protein, in contrast to a heterozygous genetic knockout of the target. Intrabodies could in this way markedly accelerate the generation of mouse models.
Applications in Oncology
A phase I clinical trial was performed to assess the feasibility of adenovirus-mediated intrabody gene therapy [271], and various strategies have been suggested for the intracellular use of antibodies to target processes related to cancer (Table 5).
Applications in Oncology: Targeting at the Cellular Level
At the cellular level, intrabodies have been used to interfere with protein–protein interactions in oncogenic pathways [272] and to influence processes involved in proliferation, the cell cycle and apoptosis. DNA repair and genomic stability were studied by a nanobody fused to a fluorescent protein that allowed tracking of poly (ADP-ribose) polymerase 1 (PARP1), a target for cancer therapy to inhibit DNA repair for sensitizing tumour cells to radio- or chemotherapy [273]. Several proof-of-concept studies have demonstrated downregulation of human epidermal growth factor receptor-2 (HER2) or inhibition of cell proliferation by intrabodies targeting HER2 [272, 274]. A nanobody directed against the tumour suppressor p53, which is involved in processes such as the cell cycle and apoptosis [275], was used to delocalize p53 by capturing it at mitochondria to generate a functional knockdown [64]. By the expression of a library of intrabodies in tumour cells, potential mechanisms by which cells are rescued from apoptosis as a contribution to cancer progression were identified [276]. The capability of intrabodies to specifically target post-translational modifications was demonstrated by an intrabody against a particular phosphorylated form of signal transducer and activator of transcription 3 (STAT3), which plays a role in various processes including proliferation and apoptosis [277]. STAT3 can be phosphorylated at different sites or remain unphosphorylated. By using an intrabody, it was possible to block the function of the tyrosine-phosphorylated form of STAT3 (pYSTAT3) but not the serine-phosphorylated form (pSSTAT3) or the unphosphorylated form (USTAT3), which is impossible for conventional small molecule-based inhibition. This was demonstrated both in vitro and in mice and illustrates the potential of intrabodies to study mechanisms based on individual functional units of proteins [278].
Applications in Oncology: Targeting at the Tissue Level
At a level superordinate to cells, processes such as adhesion, metastasis and angiogenesis have been targeted by intrabodies. As adhesion molecules, integrins have implications in metastasis [279], and, therefore, a better understanding of integrin function might be beneficial for cancer research. Integrins can form different heterodimers depending on which integrin subunits combine. The retention of a particular integrin subunit in the ER by an intrabody allowed study of how integrin heterodimers form and revealed a hierarchy in complex formation when different subunits combine [280]. Tumour invasion and metastasis were further targeted using an intrabody to block the activation of matrix metalloproteinases in an in vitro model [281, 282]. Plasma membrane extensions that also have proteolytic activity, called invadopodia, allow cancer cells to invade their surroundings and form metastases. The protein cortactin is involved in invadopodia formation, and a better understanding of this potential drug target was achieved by blocking the function of individual domains of the protein by intracellular nanobodies, which revealed different outcomes when different epitopes of cortactin were targeted [283, 284]. The GTPase RHOB is also involved in invasion. A conformation-specific intrabody that was targeted for degradation allowed selective degradation of only the guanosine triphosphate (GTP)-bound form of the protein. Targeted degradation with this conformation-specific intrabody revealed the processes of invasion and genomic instability to be associated with only one but not the other conformation of RHOB [132]. These examples impressively illustrate how intrabodies allow research on the functional consequences of complex formation in the presence of different subunits that complex in different combinations, blocking individual protein domains and individual conformations of a protein. Angiogenesis, the process of forming new blood vessels, has been successfully blocked by ER intrabodies specific for vascular endothelial growth factor receptor 2 (VEGFR2) in in vitro models [285, 286], and a bispecific intrabody allowed inhibiting angiogenesis and tumour growth by targeting VEGFR2 and the angiopoietin receptor Tie2 [287]. An adenovirus that delivered Tie2 intrabody to mice allowed a marked reduction in vessel density and significant reductions in two types of tumours, which were xenografts derived from human Kaposi’s sarcoma or human colon carcinoma [288]. The therapeutic potential of intrabodies with respect to angiogenesis may in the future be extended by broadening the range of angiogenesis-related targets attacked by intrabodies. In addition to the strategy of starving tumours by blocking angiogenesis, the normalization of tumour vascularization has emerged as another therapeutic strategy because vascularization is expected not only to supply the tumour with nutrients but also to allow the supply of chemotherapeutics, making chemotherapy more effective [289].
Applications in Oncology: Targeting Neo-Antigens
Neo-antigens or oncoproteins, aberrant proteins that exist only in tumour cells, are particularly interesting targets because they allow the often difficult discrimination of tumour and healthy cells during therapy. The oncoprotein Bcr-Abl originates from the fusion of two genes due to the formation of an aberrant chromosome, which results in a constitutively active kinase associated with chronic myelogenous leukaemia [290]. Because intrabodies can be used to de-localize proteins, an intrabody was employed to re-route Bcr-Abl to the nucleus, where it causes apoptosis. To obtain Bcr-Abl-specific intrabodies that are functional in the cytosolic milieu, IACT was employed for intrabody selection [291]. The protein Ras, which is involved in signal transduction and is frequently mutated in cancer, can become permanently activated due to mutations and influence cell growth and survival [292]. Intrabodies targeting Ras were already developed in the early 1990s by Cattaneo and colleagues [293, 294]. In a new concept, antibody–antigen interaction-dependent apoptosis (AIDA), an intrabody specific to mutant Ras, was split into two parts variable heavy chain (VH) and variable light chain (VL). Each part was fused to inactive procaspase-3 and auto-activation of caspase-3. The formation of a tri-molecular complex of mutant Ras and the two procaspase-3-fused intrabody halves allowed the induction of apoptosis upon the binding of mutant Ras. This method could be further extended to any neo-antigen for which a specific intrabody is available [295]. A different approach was taken by using intrabodies for target validation to block protein–protein interactions in disease models and subsequently find small molecules that overlap with the antibody binding site. An anti-mutant Ras intrabody allowed the identification of compounds from a small-molecule library that are able to mimic the effect of the intrabody on protein–protein interactions [296].
Applications in Oncology: Targeting Tumours Using Cellular Immunotherapy
There are recently approved cellular therapies for haematological diseases based on immune cells that are directed towards killing tumour cells [297]. The direction of immune cells as weapons against tumour cells is achieved by means of chimeric antigen receptors (CARs) on T cells or natural killer (NK) cells. CARs are expressed after ex vivo genetic engineering of cells, which are subsequently reinfused into patients [298, 299]. Ex vivo genetic engineering of lymphocytes for cellular therapy has recently also been pursued for intrabodies. As a target for CAR T cell therapy, CD7 was chosen due to its consistent expression in T cell acute lymphoblastic leukaemia. However, CAR T cells expressed CD7 as well, which resulted in killing of not only tumour cells but also of CAR T cells. To prevent this, an ER intrabody was employed to deplete expression of CD7 in CAR T cells. The expression of the ER intrabody prevented self-attack by CAR T cells and allowed them to kill only leukaemic cells [44].
Summary and Comparison of all Strategies
The expression of intrabodies in the cytosol or nucleus usually requires an involved selection procedure, but technologies have been developed to address this requirement and have allowed the generation of cytosolic intrabodies for many research areas [89, 95]. The generation of ER intrabodies is relatively straightforward, allowing more freedom in the choice of the epitope, and the folding properties of normal antibodies are sufficient. ER intrabodies have so far been found to be well-tolerated by cells and in mice. Even substantial overexpression of an ER intrabody specific for the neuronal receptor p75NTR was not found to induce ER stress, as indicated by the absence of induction of the unfolded protein response (UPR) [300], and no marked ER stress was found by other researchers examining the retention of Z alpha antitrypsin by an ER intrabody [301]. Knockdown of VCAM1 by an ER intrabody was also detected in living mice [302], and expression of the ER intrabody by transgenic mice was well-tolerated [57]. The delivery of antibodies to the cytosol as proteins is no longer a challenge in vitro but reliably applicable with standard laboratory equipment (see also Table 3). Whether there is therapeutic potential for protein delivery still needs to be further explored. Delivering proteins to endosomes or lysosomes is less challenging but may raise the question of what therapeutic effect could be obtained by targeting these compartments, especially because the delivered protein is degraded in the lysosome. A particularly interesting future application of delivering antibodies to lysosomes could be in the context of intracellular pathogens that inhibit lysosomal acidification.
In addition to the possibility of inhibiting POIs by neutralization or re-localizing POIs, all methods based on intrabody expression or antibody delivery to the cytosol allow the targeting of POIs for degradation (Table 6). Intrabodies have already been delivered by gene therapy in preclinical animal models. The delivery of proteins to lysosomes is readily achievable even in vivo and has been used for drugs that have already been approved for therapy, including an immunotoxin [55] and ADCs [303–306].
Table 6.
Comparison of strategies to target the inside of cells with antibodies
| Properties | Intrabody expression | Antibody delivery (as a protein) | ||
|---|---|---|---|---|
| Cytosol/nucleus | ER/mitochondria | Cytosol/nucleus | Other compartments | |
| Correct antibody folding possible? | ? | + | + | + |
| Epitope choice: can non-neutralizing antibodies be used? |
– (+ if combined with targeted degradation) |
+ |
– (+ if combined with targeted degradation) |
– (+ if combined with targeted degradation) |
| Antibody-mediated functional knockdown | + | + | + | ? |
| Targeted degradation? | Tags targeting for proteasomal degradation | ER: targeting for ERAD | TRIM21 for antibodies with Fc part, tags targeting for proteasomal degradation | Lysosomal targeting (e.g. LYTACs) |
| Examples of special features targeted | PTMs, individual domains, conformations | Complex formation, knockdown in selected cellular compartment | Long half-life protein degraded, tracking of unmodified endogenous POIs | |
| Deliverable in therapy? | Adoptive cell therapy, gene therapy | Adoptive cell therapy, gene therapy |
Adoptive cell therapy? Local tissue electroporation? |
+ (ADCs, approved for therapy) |
ADCs antibody–drug conjugates, ER endoplasmic reticulum, ERAD ER-associated degradation, LYTACs lysosome-targeting chimaeras, POIs proteins of interest, PTM post-translational modification, TRIM21 tripartite motif containing-21,? indicates to be determined, – indicates no, + indicates yes
The Future: Key Preclinical and Clinical Challenges
Using intrabodies for therapy is interdisciplinary, requiring expertise in intrabody-related technologies based on molecular and cellular sciences, in specific disease areas and in clinical application. This may in some cases pose a higher threshold to using this approach. However, for certain applications, there might be no suitable or feasible alternatives [8, 307]. In previous decades, the laborious and still error-prone technologies for antibody generation have complicated the use of intrabodies. This former hurdle may be responsible for the comparably late increase in research articles in this field. Antibody generation technologies and selection technologies for cytosolic intrabodies are no longer limitations. A large number of well-validated and sequence-defined antibodies are ready for use as intrabodies and delivery as proteins into cells.
There are also a variety of successful strategies to degrade a POI. A limited number of small-molecule reagents are available to bind POIs, and targeting protein–protein interactions has been reported to be challenging for small molecules, which are small and have little contact area with the protein [12–15]. Targeted degradation by protein binders would therefore have enormous potential, but therapeutic application will critically depend on the feasibility of in vivo protein delivery. Local in vivo electroporation delivery of antibodies has been performed in rats [37], and in vivo electroporation has been applied in further model organisms, e.g. locally to muscle tissue [308]. In addition to in vivo delivery to tissues, ex vivo delivery for cell therapy could be explored as an application for protein delivery in the future. For research purposes, delivering antibodies to the cytosol might become a standard method in many research areas, depending on the availability of reagents that are affordable and suitable for this application.
The application of intrabodies in clinical use depends on in vivo or ex vivo gene therapy. Particularly in the area of neurodegenerative diseases, there are comparably many preclinical in vivo studies, probably due to the urgent necessity of targeting pathologic proteins. Pathologic mechanisms associated with protein conformation are more amenable to being targeted by antibodies than by classic small-molecule therapy, which works better for inhibiting enzymes. Gene therapy to correct the gene for mutant huntingtin (mHtt) cannot remedy pathogenic mHtt protein because it may have prion-like properties and convert other proteins to pathogenic forms. Intrabodies could solve this problem. A range of preclinical animal models have demonstrated the feasibility of intrabody gene therapy, and intrabody expression has been well-tolerated in animals [57]. This might not be surprising considering that high expression of antibodies during an immune reaction should be well-tolerated. Challenges might still arise from disease-specific pathologic mechanisms, which require a more detailed understanding of the pathologic process as well as the exact functional effects of individual intrabodies on their targets. While many intrabodies block the aggregation of pathologic proteins, the aggregation rate has also been found to be increased by an intrabody [309]. Additionally, two different intrabodies have been reported by the same authors, of which one accelerated the mutant phenotype and one rescued it [310]. In cancer research, several different Bcr-Abl-specific intrabodies with a signal sequence for nuclear translocation differed in their effect on the re-localization of Bcr-Abl to the nucleus [291]. A related observation was made by Martinelli and colleagues [311]. A better mechanistic understanding of how function is affected by binding of the antibody to its target in the cell might therefore be required to select for antibodies with the intended effects. Intrabodies targeting individual post-translational modification (PTM)s could in the future contribute further to a better understanding of processes in health and disease [312, 313]. Cellular therapy employing an ER intrabody has recently been proposed as a therapeutic approach [44]. Because it provides a solution to the challenges of intrabody delivery that has been demonstrated to be feasible in approved CAR T cell therapies [297], this approach might be a particularly promising avenue for therapeutic application of intrabodies in the future.
Acknowledgements
Open Access funding provided by Projekt DEAL.
Compliance with Ethical Standards
Conflict of interest
CZ is named as an inventor on patents and patent applications in the field of cancer immunotherapy owned by his previous institution (Georg-Speyer-Haus, Institute for Tumor Biology and Experimental Therapy, Frankfurt am Main, Germany). CZ declares that this review article was written in the absence of any other commercial or financial relationships that could be construed as a potential conflict of interest. RMÖ, JR, RM and ALJM report no conflicts of interest.
Funding
This research received no specific Grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Carter PJ, Lazar GA. Next generation antibody drugs: pursuit of the “high-hanging fruit”. Nat Rev Drug Discov. 2018;17:197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- 2.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, et al. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 3.Marschall ALJ, Zhang C, Frenzel A, Schirrmann T, Hust M, Perez F, et al. Delivery of antibodies to the cytosol: debunking the myths. mAbs. 2014;6:943–956. doi: 10.4161/mabs.29268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marschall ALJ, Zhang C, Dubel S. Evaluating the delivery of proteins to the cytosol of mammalian cells. Methods Mol Biol. 2017;1513:201–208. doi: 10.1007/978-1-4939-6539-7_14. [DOI] [PubMed] [Google Scholar]
- 5.Banaszynski LA, Chen L-C, Maynard-Smith LA, Ooi AGL, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth S, Fulcher LJ, Sapkota GP. Advances in targeted degradation of endogenous proteins. Cell Mol Life Sci CMLS. 2019;76:2761–2777. doi: 10.1007/s00018-019-03112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Brolosy MA, Stainier DYR. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, et al. A method for the acute and rapid degradation of endogenous proteins. Cell. 2017;171(1692–1706):e18. doi: 10.1016/j.cell.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato Kaneko M, Ogasawara S, Kato Y. Establishment of a multi-specific monoclonal antibody MsMab-1 recognizing both IDH1 and IDH2 mutations. Tohoku J Exp Med. 2013;230:103–109. doi: 10.1620/tjem.230.103. [DOI] [PubMed] [Google Scholar]
- 10.Nowicki S, Gottlieb E. Oncometabolites: tailoring our genes. FEBS J. 2015;282:2796–2805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 12.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov AA, Khuri FR, Fu H. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol Sci. 2013;34:393–400. doi: 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higueruelo AP, Jubb H, Blundell TL. Protein-protein interactions as druggable targets: recent technological advances. Curr Opin Pharmacol. 2013;13:791–796. doi: 10.1016/j.coph.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Trenevska I, Li D, Banham AH. Therapeutic antibodies against intracellular tumor antigens. Front Immunol. 2017;8:1001. doi: 10.3389/fimmu.2017.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmansa S, Hamaratoglu F, Affolter M, Caussinus E. Dpp spreading is required for medial but not for lateral wing disc growth. Nature. 2015;527:317–322. doi: 10.1038/nature15712. [DOI] [PubMed] [Google Scholar]
- 17.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteom. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 19.Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun. 2013;4:2660. doi: 10.1038/ncomms3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller B-M, Maier J, Secker K-A, Egetemaier S-M, Parfyonova Y, Rothbauer U, et al. Chromobodies to quantify changes of endogenous protein concentration in living cells. Mol Cell Proteom. 2018;17:2518–2533. doi: 10.1074/mcp.TIR118.000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller B-M, Maier J, Weldle M, Segan S, Traenkle B, Rothbauer U. A strategy to optimize the generation of stable chromobody cell lines for visualization and quantification of endogenous proteins in living cells. Antibodies (Basel). 2019;8:E10. doi: 10.3390/antib8010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marschall ALJ, Dübel S, Böldicke T. Specific in vivo knockdown of protein function by intrabodies. mAbs. 2015;7:1010–1035. doi: 10.1080/19420862.2015.1076601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marschall ALJ, Frenzel A, Schirrmann T, Schungel M, Dubel S. Targeting antibodies to the cytoplasm. mAbs. 2011;3:3–16. doi: 10.4161/mabs.3.1.14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenzel A, Kugler J, Helmsing S, Meier D, Schirrmann T, Hust M, et al. Designing human antibodies by phage display. Transfus Med Hemother. 2017;44:312–318. doi: 10.1159/000479633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehan J, Marasco WA. Phage and yeast display. Microbiol Spectr. 2015;3:1692–1706. doi: 10.1128/microbiolspec.AID-0028-2014. [DOI] [PubMed] [Google Scholar]
- 26.Geyer CR, McCafferty J, Dubel S, Bradbury ARM, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods Mol Biol. 2012;901:11–32. doi: 10.1007/978-1-61779-931-0_2. [DOI] [PubMed] [Google Scholar]
- 27.He M, Taussig MJ. Ribosome display: cell-free protein display technology. Brief Funct Genom Proteom. 2002;1:204–212. doi: 10.1093/bfgp/1.2.204. [DOI] [PubMed] [Google Scholar]
- 28.Lofblom J. Bacterial display in combinatorial protein engineering. Biotechnol J. 2011;6:1115–1129. doi: 10.1002/biot.201100129. [DOI] [PubMed] [Google Scholar]
- 29.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konning D, Zielonka S, Grzeschik J, Empting M, Valldorf B, Krah S, et al. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr Opin Struct Biol. 2017;45:10–16. doi: 10.1016/j.sbi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LGJ, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu JL, Zabetakis D, Brown JC, Anderson GP, Goldman ER. Thermal stability and refolding capability of shark derived single domain antibodies. Mol Immunol. 2014;59:194–199. doi: 10.1016/j.molimm.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Stahl S, Graslund T, Eriksson Karlstrom A, Frejd FY, Nygren P-A, Lofblom J. Affibody molecules in biotechnological and medical applications. Trends Biotechnol. 2017;35:691–712. doi: 10.1016/j.tibtech.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Valerio-Lepiniec M, Urvoas A, Chevrel A, Guellouz A, Ferrandez Y, Mesneau A, et al. The alphaRep artificial repeat protein scaffold: a new tool for crystallization and live cell applications. Biochem Soc Trans. 2015;43:819–824. doi: 10.1042/BST20150075. [DOI] [PubMed] [Google Scholar]
- 35.Pluckthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu Rev Pharmacol Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 36.Koide A, Koide S. Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- 37.Lan C-Y, Tan P-H, Cheng J-T, Lu H-F, Lin M-W, Hsiao P-N, et al. Immunoneutralization of c-Fos using intrathecal antibody electroporation attenuates chronic constrictive injury-induced hyperalgesia and regulates preprodynorphin expression in rats. Anesthesiology. 2003;99:938–946. doi: 10.1097/00000542-200310000-00029. [DOI] [PubMed] [Google Scholar]
- 38.ClinicalTrials.gov. Search result: gene therapy. https://clinicaltrials.gov/ct2/results?cond=%26term=%26type=%26rslt=%26age_v=%26gndr=%26intr=Gene+therapy+%26titles=%26outc=%26spons=%26lead=%26id=%26cntry=%26state=%26city=%26dist=%26locn=%26rsub=%26strd_s=%26strd_e=%26prcd_s=%26prcd_e=%26sfpd_s=%26sfpd_e=%26rfpd_s=%26rfpd_e=%26lupd_s=%26lupd_e=%26sort. Accessed 25 Mar 2020.
- 39.US FDA. Approved cellular and gene therapy products. https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/approved-cellular-and-gene-therapy-products. Accessed 25 Mar 2020.
- 40.US FDA. IMLYGIC (talimogene laherparepvec). https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/imlygic-talimogene-laherparepvec. Accessed 25 Mar 2020.
- 41.US FDA. Zolgensma. https://www.fda.gov/vaccines-blood-biologics/zolgensma. Accessed 25 Mar 2020.
- 42.US FDA. Luxturna. https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/luxturna. Accessed 25 Mar 2020.
- 43.Keeler A, ElMallah M, Flotte T. Gene therapy 2017: progress and future directions. Clin Transl Sci. 2017;10:242–248. doi: 10.1111/cts.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Png YT, Vinanica N, Kamiya T, Shimasaki N, Coustan-Smith E, Campana D. Blockade of CD7 expression in T cells for effective chimeric antigen receptor targeting of T-cell malignancies. Blood Adv. 2017;1:2348–2360. doi: 10.1182/bloodadvances.2017009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.US FDA. Kymriah (tisagenlecleucel). https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/kymriah-tisagenlecleucel. Accessed 25 Mar 2020.
- 46.US FDA. YESCARTA (axicabtagene ciloleucel). https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/yescarta-axicabtagene-ciloleucel
- 47.European Medicines Agency. Strimvelis. EMEA/H/C/003854. https://www.ema.europa.eu/en/medicines/human/EPAR/strimvelis. Accessed 25 Mar 2020.
- 48.Kaufmann KB, Buning H, Galy A, Schambach A, Grez M. Gene therapy on the move. EMBO Mol Med. 2013;5:1642–1661. doi: 10.1002/emmm.201202287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papapetrou EP, Schambach A. Gene insertion into genomic safe harbors for human gene therapy. Mol Ther. 2016;24:678–684. doi: 10.1038/mt.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anguela XM, High KA. Entering the modern era of gene therapy. Annu Rev Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 51.Biocca S, Ruberti F, Tafani M, Pierandrei-Amaldi P, Cattaneo A. Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Biotechnology (Nat Publ Co) 1995;13:1110–1115. doi: 10.1038/nbt1095-1110. [DOI] [PubMed] [Google Scholar]
- 52.Biocca S, Cattaneo A. Intracellular immunization: antibody targeting to subcellular compartments. Trends Cell Biol. 1995;5:248–252. doi: 10.1016/s0962-8924(00)89019-4. [DOI] [PubMed] [Google Scholar]
- 53.Cardinale A, Biocca S. Expressing intrabodies in mammalian cells. In: Kontermann R, Dübel S, editors. Antibody engineering. Berlin: Springer; 2010. pp. 161–172. [Google Scholar]
- 54.Beck A, Goetsch L, Dumontet C, Corvaia N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat Rev Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 55.Dhillon S. Moxetumomab pasudotox: first global approval. Drugs. 2018;78:1763–1767. doi: 10.1007/s40265-018-1000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strebe N, Guse A, Schüngel M, Schirrmann T, Hafner M, Jostock T, et al. Functional knockdown of VCAM-1 at the posttranslational level with ER retained antibodies. J Immunol Methods. 2009;341:30–40. doi: 10.1016/j.jim.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 57.Marschall ALJ, Single FN, Schlarmann K, Bosio A, Strebe N, van den Heuvel J, et al. Functional knock down of VCAM1 in mice mediated by endoplasmatic reticulum retained intrabodies. mAbs. 2014;6:1394–1401. doi: 10.4161/mabs.34377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Büssow K, Themann P, Luu S, Pentrowski P, Harting C, Majewski M, et al. ER intrabody-mediated inhibition of interferon alpha secretion by mouse macrophages and dendritic cells. PLoS One. 2019;14:e0215062. doi: 10.1371/journal.pone.0215062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marschall ALJ, Dübel S. Antibodies inside of a cell can change its outside: can intrabodies provide a new therapeutic paradigm? Comput Struct Biotechnol J. 2016;14:304–308. doi: 10.1016/j.csbj.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marschall ALJ, Dubel S, Boldicke T. Recent advances with ER targeted intrabodies. Adv Exp Med Biol. 2015;917:77–93. doi: 10.1007/978-3-319-32805-8_5. [DOI] [PubMed] [Google Scholar]
- 61.Traub LM. A nanobody-based molecular toolkit provides new mechanistic insight into clathrin-coat initiation. eLife. 2019;8:e41768. doi: 10.7554/eLife.41768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prole DL, Taylor CW. A genetically encoded toolkit of functionalized nanobodies against fluorescent proteins for visualizing and manipulating intracellular signalling. BMC Biol. 2019;17:41. doi: 10.1186/s12915-019-0662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beghein E, Van Audenhove I, Zwaenepoel O, Verhelle A, De Ganck A, Gettemans J. A new survivin tracer tracks, delocalizes and captures endogenous survivin at different subcellular locations and in distinct organelles. Sci Rep. 2016;6:31177. doi: 10.1038/srep31177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steels A, Verhelle A, Zwaenepoel O, Gettemans J. Intracellular displacement of p53 using transactivation domain (p53 TAD) specific nanobodies. mAbs. 2018;10:1045–1059. doi: 10.1080/19420862.2018.1502025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moody PR, Sayers EJ, Magnusson JP, Alexander C, Borri P, Watson P, et al. Receptor crosslinking: a general method to trigger internalization and lysosomal targeting of therapeutic receptor:ligand complexes. Mol Ther J. 2015;23:1888–1898. doi: 10.1038/mt.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel SG, Sayers EJ, He L, Narayan R, Williams TL, Mills EM, et al. Cell-penetrating peptide sequence and modification dependent uptake and subcellular distribution of green florescent protein in different cell lines. Sci Rep. 2019;9:6298. doi: 10.1038/s41598-019-42456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang RE, Liu T, Wang Y, Cao Y, Du J, Luo X, et al. An immunosuppressive antibody–drug conjugate. J Am Chem Soc. 2015;137:3229–3232. doi: 10.1021/jacs.5b00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, et al. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015;527:323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 69.Lambert JM, Chari RVJ. Ado-trastuzumab Emtansine (T-DM1): an antibody–drug conjugate (ADC) J Med Chem. 2014;57:6949–6964. doi: 10.1021/jm500766w. [DOI] [PubMed] [Google Scholar]
- 70.Erickson HK, Park PU, Widdison WC, Kovtun YV, Garrett LM, Hoffman K, et al. Antibody-maytansinoid conjugates are activated in targeted cancer cells by lysosomal degradation and linker-dependent intracellular processing. Cancer Res. 2006;66:4426–4433. doi: 10.1158/0008-5472.CAN-05-4489. [DOI] [PubMed] [Google Scholar]
- 71.Lambert JM, Morris CQ. Antibody–drug conjugates (ADCs) for personalized treatment of solid tumors: a review. Adv Ther. 2017;34:1015–1035. doi: 10.1007/s12325-017-0519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drake PM, Rabuka D. Recent developments in ADC technology: preclinical studies signal future clinical trends. BioDrugs. 2017;31:521–531. doi: 10.1007/s40259-017-0254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rust A, Partridge LJ, Davletov B, Hautbergue GM. The use of plant-derived ribosome inactivating proteins in immunotoxin development: past, present and future generations. Toxins. 2017;9:344. doi: 10.3390/toxins9110344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dosio F, Arpicco S, Adobati E, Canevari S, Brusa P, De Santis R, et al. Role of cross-linking agents in determining the biochemical and pharmacokinetic properties of Mgr6-clavin immunotoxins. Bioconjug Chem. 1998;9:372–381. doi: 10.1021/bc970192w. [DOI] [PubMed] [Google Scholar]
- 75.Weldon JE, Skarzynski M, Therres JA, Ostovitz JR, Zhou H, Kreitman RJ, et al. Designing the furin-cleavable linker in recombinant immunotoxins based on Pseudomonas exotoxin A. Bioconjug Chem. 2015;26:1120–1128. doi: 10.1021/acs.bioconjchem.5b00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lord JM, Deeks E, Marsden CJ, Moore K, Pateman C, Smith DC, et al. Retrograde transport of toxins across the endoplasmic reticulum membrane. Biochem Soc Trans. 2003;31:1260–1262. doi: 10.1042/bst0311260. [DOI] [PubMed] [Google Scholar]
- 77.Simpson JC, Roberts LM, Romisch K, Davey J, Wolf DH, Lord JM. Ricin A chain utilises the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett. 1999;459:80–84. doi: 10.1016/s0014-5793(99)01222-3. [DOI] [PubMed] [Google Scholar]
- 78.Deeks ED, Cook JP, Day PJ, Smith DC, Roberts LM, Lord JM. The low lysine content of ricin A chain reduces the risk of proteolytic degradation after translocation from the endoplasmic reticulum to the cytosol. Biochemistry. 2002;41:3405–3413. doi: 10.1021/bi011580v. [DOI] [PubMed] [Google Scholar]
- 79.Spooner RA, Hart PJ, Cook JP, Pietroni P, Rogon C, Höhfeld J, et al. Cytosolic chaperones influence the fate of a toxin dislocated from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2008;105:17408–17413. doi: 10.1073/pnas.0809013105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alewine C, Hassan R, Pastan I. Advances in anticancer immunotoxin therapy. Oncologist. 2015;20:176–185. doi: 10.1634/theoncologist.2014-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sehnert B, Burkhardt H, Wessels JT, Schroder A, May MJ, Vestweber D, et al. NF-kappaB inhibitor targeted to activated endothelium demonstrates a critical role of endothelial NF-kappaB in immune-mediated diseases. Proc Natl Acad Sci USA. 2013;110:16556–16561. doi: 10.1073/pnas.1218219110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sehnert B, Burkhardt H, Dubel S, Voll RE. The “sneaking-ligand” approach: cell-type specific inhibition of the classical. Methods Mol Biol. 2015;1280:559–578. doi: 10.1007/978-1-4939-2422-6_33. [DOI] [PubMed] [Google Scholar]
- 83.der Maur AA, Tissot K, Barberis A. Antigen-independent selection of intracellular stable antibody frameworks. Methods. 2004;34:215–224. doi: 10.1016/j.ymeth.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 84.Visintin M, Quondam M, Cattaneo A. The intracellular antibody capture technology: towards the high-throughput selection of functional intracellular antibodies for target validation. Methods. 2004;34:200–214. doi: 10.1016/j.ymeth.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 85.Kabayama H, Takeuchi M, Tokushige N, Muramatsu S, Kabayama M, Fukuda M, et al. An ultra-stable cytoplasmic antibody engineered for in vivo applications. Nat Commun. 2020;11:336. doi: 10.1038/s41467-019-13654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanaka T, Rabbitts TH. Intrabodies based on intracellular capture frameworks that bind the RAS protein with high affinity and impair oncogenic transformation. EMBO J. 2003;22:1025–1035. doi: 10.1093/emboj/cdg106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.da Silva FA, Santa-Marta M, Freitas-Vieira A, Mascarenhas P, Barahona I, Moniz-Pereira J, et al. Camelized rabbit-derived VH single-domain intrabodies against Vif strongly neutralize HIV-1 infectivity. J Mol Biol. 2004;340:525–542. doi: 10.1016/j.jmb.2004.04.062. [DOI] [PubMed] [Google Scholar]
- 88.Saerens D, Pellis M, Loris R, Pardon E, Dumoulin M, Matagne A, et al. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J Mol Biol. 2005;352:597–607. doi: 10.1016/j.jmb.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 89.Visintin M, Settanni G, Maritan A, Graziosi S, Marks JD, Cattaneo A. The intracellular antibody capture technology (IACT): towards a consensus sequence for intracellular antibodies. J Mol Biol. 2002;317:73–83. doi: 10.1006/jmbi.2002.5392. [DOI] [PubMed] [Google Scholar]
- 90.Tanaka T, Rabbitts TH. Functional intracellular antibody fragments do not require invariant intra-domain disulfide bonds. J Mol Biol. 2008;376:749–757. doi: 10.1016/j.jmb.2007.11.085. [DOI] [PubMed] [Google Scholar]
- 91.Boldicke T. Single domain antibodies for the knockdown of cytosolic and nuclear proteins. Protein Sci. 2017;26:925–945. doi: 10.1002/pro.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Woods J. Selection of functional intracellular nanobodies. SLAS Discov. 2019;24:703–713. doi: 10.1177/2472555219853235. [DOI] [PubMed] [Google Scholar]
- 93.Kvam E, Sierks MR, Shoemaker CB, Messer A. Physico-chemical determinants of soluble intrabody expression in mammalian cell cytoplasm. Protein Eng Des Sel PEDS. 2010;23:489–498. doi: 10.1093/protein/gzq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gross GG, Junge JA, Mora RJ, Kwon H-B, Olson CA, Takahashi TT, et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fisher AC, DeLisa MP. Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery. J Mol Biol. 2009;385:299–311. doi: 10.1016/j.jmb.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pellis M, Pardon E, Zolghadr K, Rothbauer U, Vincke C, Kinne J, et al. A bacterial-two-hybrid selection system for one-step isolation of intracellularly functional Nanobodies. Arch Biochem Biophys. 2012;526:114–123. doi: 10.1016/j.abb.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 97.Nguyen TD, Nagamune T, Kawahara M. A suicide switch directly eliminates intracellular scFv oligomers in the cytoplasm of mammalian cells. Biotechnol J. 2019;14:e1800350. doi: 10.1002/biot.201800350. [DOI] [PubMed] [Google Scholar]
- 98.Siciliano V, DiAndreth B, Monel B, Beal J, Huh J, Clayton KL, et al. Engineering modular intracellular protein sensor-actuator devices. Nat Commun. 2018;9:1881. doi: 10.1038/s41467-018-03984-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Verdurmen WPR, Mazlami M, Plückthun A. A quantitative comparison of cytosolic delivery via different protein uptake systems. Sci Rep. 2017;7:13194. doi: 10.1038/s41598-017-13469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stewart MP, Sharei A, Ding X, Sahay G, Langer R, Jensen KF. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 101.El-Sayed A, Futaki S, Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pei D, Buyanova M. Overcoming endosomal entrapment in drug delivery. Bioconjug Chem. 2019;30:273–283. doi: 10.1021/acs.bioconjchem.8b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu J, Xiong R, Brans T, Lippens S, Parthoens E, Zanacchi FC, et al. Repeated photoporation with graphene quantum dots enables homogeneous labeling of live cells with extrinsic markers for fluorescence microscopy. Light Sci Appl. 2018;7:47. doi: 10.1038/s41377-018-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blanco-Toribio A, Muyldermans S, Frankel G, Fernandez LA. Direct injection of functional single-domain antibodies from E. coli into human cells. PloS One. 2010;5:e15227. [DOI] [PMC free article] [PubMed]
- 105.Freund G, Sibler A-P, Desplancq D, Oulad-Abdelghani M, Vigneron M, Gannon J, et al. Targeting endogenous nuclear antigens by electrotransfer of monoclonal antibodies in living cells. mAbs. 2013;5:518–522. doi: 10.4161/mabs.25084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stewart MP, Langer R, Jensen KF. Intracellular delivery by membrane disruption: mechanisms, strategies, and concepts. Chem Rev. 2018;118:7409–7531. doi: 10.1021/acs.chemrev.7b00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chakrabarti R, Wylie DE, Schuster SM. Transfer of monoclonal antibodies into mammalian cells by electroporation. J Biol Chem. 1989;264:15494–15500. [PubMed] [Google Scholar]
- 108.Conic S, Desplancq D, Ferrand A, Fischer V, Heyer V, Reina S, Martin B, et al. Imaging of native transcription factors and histone phosphorylation at high resolution in live cells. J Cell Biol. 2018;217:1537–1552. doi: 10.1083/jcb.201709153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 110.Chung HK, Jacobs CL, Huo Y, Yang J, Krumm SA, Plemper RK, et al. Tunable and reversible drug control of protein production via a self-excising degron. Nat Chem Biol. 2015;11:713–720. doi: 10.1038/nchembio.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Navarro R, Chen L-C, Rakhit R, Wandless TJ. A novel destabilizing domain based on a small-molecule dependent fluorophore. ACS Chem Biol. 2016;11:2101–2104. doi: 10.1021/acschembio.6b00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mullard A. First targeted protein degrader hits the clinic. Nat Rev Drug Discov. 2019 doi: 10.1038/d41573-019-00043-6. [DOI] [PubMed] [Google Scholar]
- 113.Pettersson M, Crews CM. PROteolysis TArgeting Chimeras (PROTACs)—past, present and future. Drug Discov Today Technol. 2019;31:15–27. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discov Today. 2015;20:838–847. doi: 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 115.Zou Y, Ma D, Wang Y. The PROTAC technology in drug development. Cell Biochem Funct. 2019;37:21–30. doi: 10.1002/cbf.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Caruso C. Arvinas, Pfizer team up on PROTACs. Cancer Discov. 2018;8:377–378. doi: 10.1158/2159-8290.CD-NB2018-015. [DOI] [PubMed] [Google Scholar]
- 117.Guo J, Liu J, Wei W. Degrading proteins in animals: “PROTAC”tion goes in vivo. Cell Res. 2019;29:179–180. doi: 10.1038/s41422-019-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arvinas Inc. a phase 1 clinical trial of ARV-110 in patients with metastatic castration-resistant prostate cancer (mCRPC) [ClinicalTrials.gov identifier NCT03888612]. National Institutes of Health, ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03888612. Accessed 25 Mar 2020.
- 119.Burbelo PD, Ching KH, Han BL, Bush ER, Reeves WH, Iadarola MJ. Extraordinary antigenicity of the human Ro52 autoantigen. Am J Transl Res. 2010;2:145–155. [PMC free article] [PubMed] [Google Scholar]
- 120.Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci USA. 2010;107:19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, et al. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–2516. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burkhardt S, Borsos M, Szydlowska A, Godwin J, Williams SA, Cohen PE, et al. Chromosome cohesion established by Rec8-cohesin in fetal oocytes is maintained without detectable turnover in oocytes arrested for months in mice. Curr Biol. 2016;26:678–685. doi: 10.1016/j.cub.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Caussinus E, Kanca O, Affolter M. Protein knockouts in living eukaryotes using deGradFP and green fluorescent protein fusion targets. Curr Protoc Protein Sci. 2013;73:30–32. doi: 10.1002/0471140864.ps3002s73. [DOI] [PubMed] [Google Scholar]
- 124.Ochoa-Espinosa A, Harmansa S, Caussinus E, Affolter M. Myosin II is not required for Drosophila tracheal branch elongation and cell intercalation. Development. 2017;144:2961–2968. doi: 10.1242/dev.148940. [DOI] [PubMed] [Google Scholar]
- 125.Harmansa S, Affolter M. Protein binders and their applications in developmental biology. Development. 2018;145:dev148874. doi: 10.1242/dev.148874. [DOI] [PubMed] [Google Scholar]
- 126.Yamaguchi N, Colak-Champollion T, Knaut H. zGrad is a nanobody-based degron system that inactivates proteins in zebrafish. eLife. 2019;8:e43125. doi: 10.7554/eLife.43125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Portnoff AD, Stephens EA, Varner JD, DeLisa MP. Ubiquibodies, synthetic E3 ubiquitin ligases endowed with unnatural substrate specificity for targeted protein silencing. J Biol Chem. 2014;289:7844–7855. doi: 10.1074/jbc.M113.544825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shin YJ, Park SK, Jung YJ, Kim YN, Kim KS, Park OK, et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci Rep. 2015;5:14269. doi: 10.1038/srep14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fulcher LJ, Hutchinson LD, Macartney TJ, Turnbull C, Sapkota GP. Targeting endogenous proteins for degradation through the affinity-directed protein missile system. Open Biol. 2017;7:170066. doi: 10.1098/rsob.170066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chatterjee D, Bhatt M, Butler D, De Genst E, Dobson CM, Messer A, et al. Proteasome-targeted nanobodies alleviate pathology and functional decline in an alpha-synuclein-based Parkinson’s disease model. NPJ Parkinsons Dis. 2018;4:25. doi: 10.1038/s41531-018-0062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Daniel K, Icha J, Horenburg C, Muller D, Norden C, Mansfeld J. Conditional control of fluorescent protein degradation by an auxin-dependent nanobody. Nat Commun. 2018;9:3297. doi: 10.1038/s41467-018-05855-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bery N, Keller L, Soulie M, Gence R, Iscache A-L, Cherier J, et al. A targeted protein degradation cell-based screening for nanobodies selective toward the cellular RHOB GTP-bound conformation. Cell Chem Biol. 2019;26(1544–1558):e6. doi: 10.1016/j.chembiol.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 133.Butler DC, Messer A. Bifunctional anti-huntingtin proteasome-directed intrabodies mediate efficient degradation of mutant huntingtin exon 1 protein fragments. PLoS One. 2011;6:e29199. doi: 10.1371/journal.pone.0029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tamaki Y, Shodai A, Morimura T, Hikiami R, Minamiyama S, Ayaki T, et al. Elimination of TDP-43 inclusions linked to amyotrophic lateral sclerosis by a misfolding-specific intrabody with dual proteolytic signals. Sci Rep. 2018;8:6030. doi: 10.1038/s41598-018-24463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Banik S, Pedram K, Wisnovsky S, Riley N, Bertozzi C. Lysosome targeting chimeras (LYTACs) for the degradation of secreted and membrane proteins. ChemRxiv Preprint. 2019. https://chemrxiv.org/articles/Lysosome_Targeting_Chimeras_LYTACs_for_the_Degradation_of_Secreted_and_Membrane_Proteins/7927061/1. Accessed 17 Mar 2020.
- 136.Vecchi L, Petris G, Bestagno M, Burrone OR. Selective targeting of proteins within secretory pathway for endoplasmic reticulum-associated degradation. J Biol Chem. 2012;287:20007–20015. doi: 10.1074/jbc.M112.355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 138.Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PubMed] [Google Scholar]
- 139.Delenclos M, Burgess JD, Lamprokostopoulou A, Outeiro TF, Vekrellis K, McLean PJ. Cellular models of alpha-synuclein toxicity and aggregation. J Neurochem. 2019;150:566–576. doi: 10.1111/jnc.14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sonawane SK, Chinnathambi S. Prion-like propagation of post-translationally modified tau in Alzheimer’s disease: a hypothesis. J Mol Neurosci MN. 2018;65:480–490. doi: 10.1007/s12031-018-1111-5. [DOI] [PubMed] [Google Scholar]
- 141.Vasili E, Dominguez-Meijide A, Outeiro TF. Spreading of alpha-synuclein and tau: a systematic comparison of the mechanisms involved. Front Mol Neurosci. 2019;12:107. doi: 10.3389/fnmol.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Durães F, Pinto M, Sousa E. Old drugs as new treatments for neurodegenerative diseases. Pharmaceuticals (Basel). 2018;11:44. doi: 10.3390/ph11020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bosco DA, LaVoie MJ, Petsko GA, Ringe D. Proteostasis and movement disorders: Parkinson’s disease and amyotrophic lateral sclerosis. Cold Spring Harb Perspect Biol. 2011;3:a007500. doi: 10.1101/cshperspect.a007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou C, Emadi S, Sierks MR, Messer A. A human single-chain Fv intrabody blocks aberrant cellular effects of overexpressed alpha-synuclein. Mol Ther. 2004;10:1023–1031. doi: 10.1016/j.ymthe.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 145.Lynch SM, Zhou C, Messer A. An scFv intrabody against the nonamyloid component of alpha-synuclein reduces intracellular aggregation and toxicity. J Mol Biol. 2008;377:136–147. doi: 10.1016/j.jmb.2007.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yuan B, Sierks MR. Intracellular targeting and clearance of oligomeric alpha-synuclein alleviates toxicity in mammalian cells. Neurosci Lett. 2009;459:16–18. doi: 10.1016/j.neulet.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 147.Joshi SN, Butler DC, Messer A. Fusion to a highly charged proteasomal retargeting sequence increases soluble cytoplasmic expression and efficacy of diverse anti-synuclein intrabodies. mAbs. 2012;4:686–693. doi: 10.4161/mabs.21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Butler DC, Joshi SN, Genst ED, Baghel AS, Dobson CM, Messer A. Bifunctional anti-non-amyloid component alpha-synuclein nanobodies are protective in situ. PLoS One. 2016;11:e0165964. doi: 10.1371/journal.pone.0165964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cardinale A, Merlo D, Giunchedi P, Biocca S. Therapeutic application of intrabodies against age-related neurodegenerative disorders. Curr Pharm Des. 2014;20:6028–6036. doi: 10.2174/1381612820666140314121444. [DOI] [PubMed] [Google Scholar]
- 150.Meli G, Krako N, Manca A, Lecci A, Cattaneo A. Intrabodies for protein interference in Alzheimers disease. J Biol Regul Homeost Agents. 2013;27:89–105. [PubMed] [Google Scholar]
- 151.Masters CL, Cappai R, Barnham KJ, Villemagne VL. Molecular mechanisms for Alzheimer’s disease: implications for neuroimaging and therapeutics. J Neurochem. 2006;97:1700–1725. doi: 10.1111/j.1471-4159.2006.03989.x. [DOI] [PubMed] [Google Scholar]
- 152.Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. beta-site specific intrabodies to decrease and prevent generation of Alzheimer’s Abeta peptide. J Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hayashi I, Takatori S, Urano Y, Iwanari H, Isoo N, Osawa S, et al. Single chain variable fragment against nicastrin inhibits the gamma-secretase activity. J Biol Chem. 2009;284:27838–27847. doi: 10.1074/jbc.M109.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Meli G, Lecci A, Manca A, Krako N, Albertini V, Benussi L, et al. Conformational targeting of intracellular Abeta oligomers demonstrates their pathological oligomerization inside the endoplasmic reticulum. Nat Commun. 2014;5:3867. doi: 10.1038/ncomms4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sudol KL, Mastrangelo MA, Narrow WC, Frazer ME, Levites YR, Golde TE, et al. Generating differentially targeted amyloid-beta specific intrabodies as a passive vaccination strategy for Alzheimer’s disease. Mol Ther. 2009;17:2031–2040. doi: 10.1038/mt.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Desai MK, Mastrangelo MA, Ryan DA, Sudol KL, Narrow WC, Bowers WJ. Early oligodendrocyte/myelin pathology in Alzheimer’s disease mice constitutes a novel therapeutic target. Am J Pathol. 2010;177:1422–1435. doi: 10.2353/ajpath.2010.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cook SG, Goodell DJ, Restrepo S, Arnold DB, Bayer KU. Simultaneous live imaging of multiple endogenous proteins reveals a mechanism for Alzheimer’s-related plasticity impairment. Cell Rep. 2019;27(658–665):e4. doi: 10.1016/j.celrep.2019.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dong J-X, Lee Y, Kirmiz M, Palacio S, Dumitras C, Moreno CM, et al. A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. eLife. 2019;8:e48750. doi: 10.7554/eLife.48750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rojas JC, Boxer AL. Targeting tauopathies for therapeutic translation. Nat Rev Neurol. 2016;12:74–76. doi: 10.1038/nrneurol.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Messer A, Butler DC. Optimizing intracellular antibodies (intrabodies/nanobodies) to treat neurodegenerative disorders. Neurobiol Dis. 2020;134:104619. doi: 10.1016/j.nbd.2019.104619. [DOI] [PubMed] [Google Scholar]
- 161.Melchionna T, Cattaneo A. A protein silencing switch by ligand-induced proteasome-targeting intrabodies. J Mol Biol. 2007;374:641–654. doi: 10.1016/j.jmb.2007.09.053. [DOI] [PubMed] [Google Scholar]
- 162.Di Primio C, Quercioli V, Siano G, Kovacech B, Novak M, Cattaneo A. Conformational dynamics of Tau in the cell quantified by an intramolecular FRET biosensor in physiological and pathological context. bioRxiv. 2016 doi: 10.1101/041756. [DOI] [Google Scholar]
- 163.Gallardo G, Wong CH, Ricardez SM, Mann CN, Lin KH, Leyns CEG, et al. Targeting tauopathy with engineered tau-degrading intrabodies. Mol Neurodegener. 2019;14:38. doi: 10.1186/s13024-019-0340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu C, Song X, Nisbet R, Gotz J. Co-immunoprecipitation with tau isoform-specific antibodies reveals distinct protein interactions and highlights a putative role for 2 N tau in disease. J Biol Chem. 2016;291:8173–8188. doi: 10.1074/jbc.M115.641902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nisbet RM, Van der Jeugd A, Leinenga G, Evans HT, Janowicz PW, Gotz J. Combined effects of scanning ultrasound and a tau-specific single chain antibody in a tau transgenic mouse model. Brain J Neurol. 2017;140:1220–1230. doi: 10.1093/brain/awx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jimenez-Sanchez M, Licitra F, Underwood BR, Rubinsztein DC. Huntington’s disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb Perspect Med. 2017;7:a024240. doi: 10.1101/cshperspect.a024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pearce MMP, Kopito RR. Prion-like characteristics of polyglutamine-containing proteins. Cold Spring Harb Perspect Med. 2018;8:a024257. doi: 10.1101/cshperspect.a024257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Denis HL, Lauruol F, Cicchetti F. Are immunotherapies for Huntington’s disease a realistic option? Mol Psychiatry. 2019;24:364–377. doi: 10.1038/s41380-018-0021-9. [DOI] [PubMed] [Google Scholar]
- 169.Denis HL, David LS, Cicchetti F. Antibody-based therapies for Huntington’s disease: current status and future directions. Neurobiol Dis. 2019;132:104569. doi: 10.1016/j.nbd.2019.104569. [DOI] [PubMed] [Google Scholar]
- 170.Bortvedt SF, McLear JA, Messer A, Ahern-Rindell AJ, Wolfgang WJ. Cystamine and intrabody co-treatment confers additional benefits in a fly model of Huntington’s disease. Neurobiol Dis. 2010;40:130–134. doi: 10.1016/j.nbd.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Colby DW, Chu Y, Cassady JP, Duennwald M, Zazulak H, Webster JM, et al. Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody. Proc Natl Acad Sci USA. 2004;101:17616–17621. doi: 10.1073/pnas.0408134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Southwell AL, Khoshnan A, Dunn DE, Bugg CW, Lo DC, Patterson PH. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci. 2008;28:9013–9020. doi: 10.1523/JNEUROSCI.2747-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Southwell AL, Ko J, Patterson PH. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington’s disease. J Neurosci. 2009;29:13589–13602. doi: 10.1523/JNEUROSCI.4286-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Snyder-Keller A, McLear JA, Hathorn T, Messer A. Early or late-stage anti-N-terminal Huntingtin intrabody gene therapy reduces pathological features in B6.HDR6/1 mice. J Neuropathol Exp Neurol. 2010;69:1078–1085. doi: 10.1097/NEN.0b013e3181f530ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Amaro IA, Henderson LA. An intrabody drug (rAAV6-INT41) reduces the binding of N-terminal Huntingtin fragment(s) to DNA to basal levels in PC12 cells and delays cognitive loss in the R6/2 animal model. J Neurodegener Dis. 2016;2016:7120753. doi: 10.1155/2016/7120753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primer. 2017;3:17071. doi: 10.1038/nrdp.2017.71. [DOI] [PubMed] [Google Scholar]
- 177.Maurel C, Dangoumau A, Marouillat S, Brulard C, Chami A, Hergesheimer R, et al. Causative genes in amyotrophic lateral sclerosis and protein degradation pathways: a link to neurodegeneration. Mol Neurobiol. 2018;55:6480–6499. doi: 10.1007/s12035-017-0856-0. [DOI] [PubMed] [Google Scholar]
- 178.Alsultan AA, Waller R, Heath PR, Kirby J. The genetics of amyotrophic lateral sclerosis: current insights. Degener Neurol Neuromuscul Dis. 2016;6:49–64. doi: 10.2147/DNND.S84956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Millecamps S, Salachas F, Cazeneuve C, Gordon P, Bricka B, Camuzat A, et al. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J Med Genet. 2010;47:554–560. doi: 10.1136/jmg.2010.077180. [DOI] [PubMed] [Google Scholar]
- 180.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 181.Ghadge GD, Kay BK, Drigotas C, Roos RP. Single chain variable fragment antibodies directed against SOD1 ameliorate disease in mutant SOD1 transgenic mice. Neurobiol Dis. 2019;121:131–137. doi: 10.1016/j.nbd.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 182.Patel P, Kriz J, Gravel M, Soucy G, Bareil C, Gravel C, et al. Adeno-associated virus-mediated delivery of a recombinant single-chain antibody against misfolded superoxide dismutase for treatment of amyotrophic lateral sclerosis. Mol Ther. 2014;22:498–510. doi: 10.1038/mt.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mackenzie IRA, Neumann M. Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem. 2016;138(Suppl 1):54–70. doi: 10.1111/jnc.13588. [DOI] [PubMed] [Google Scholar]
- 184.Pozzi S, Thammisetty SS, Codron P, Rahimian R, Plourde KV, Soucy G, et al. Virus-mediated delivery of antibody targeting TAR DNA-binding protein-43 mitigates associated neuropathology. J Clin Investig. 2019;129:1581–1595. doi: 10.1172/JCI123931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chartier A, Raz V, Sterrenburg E, Verrips CT, van der Maarel SM, Simonelig M. Prevention of oculopharyngeal muscular dystrophy by muscular expression of Llama single-chain intrabodies in vivo. Hum Mol Genet. 2009;18:1849–1859. doi: 10.1093/hmg/ddp101. [DOI] [PubMed] [Google Scholar]
- 186.Gueorguieva D, Li S, Walsh N, Mukerji A, Tanha J, Pandey S. Identification of single-domain, Bax-specific intrabodies that confer resistance to mammalian cells against oxidative-stress-induced apoptosis. FASEB J. 2006;20:2636–2638. doi: 10.1096/fj.06-6306fje. [DOI] [PubMed] [Google Scholar]
- 187.Richardson JH, Sodroski JG, Waldmann TA, Marasco WA. Phenotypic knockout of the high-affinity human interleukin 2 receptor by intracellular single-chain antibodies against the alpha subunit of the receptor. Proc Natl Acad Sci USA. 1995;92:3137–3141. doi: 10.1073/pnas.92.8.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Richardson JH, Hofmann W, Sodroski JG, Marasco WA. Intrabody-mediated knockout of the high-affinity IL-2 receptor in primary human T cells using a bicistronic lentivirus vector. Gene Ther. 1998;5:635–644. doi: 10.1038/sj.gt.3300644. [DOI] [PubMed] [Google Scholar]
- 189.Steinberger P, Andris-Widhopf J, Buhler B, Torbett BE, Barbas CF., 3rd Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion. Proc Natl Acad Sci USA. 2000;97:805–810. doi: 10.1073/pnas.97.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.BouHamdan M, Strayer DS, Wei D, Mukhtar M, Duan LX, Hoxie J, et al. Inhibition of HIV-1 infection by down-regulation of the CXCR4 co-receptor using an intracellular single chain variable fragment against CXCR4. Gene Ther. 2001;8:408–418. doi: 10.1038/sj.gt.3301411. [DOI] [PubMed] [Google Scholar]
- 191.Cordelier P, Kulkowsky JW, Ko C, Matskevitch AA, McKee HJ, Rossi JJ, et al. Protecting from R5-tropic HIV: individual and combined effectiveness of a hammerhead ribozyme and a single-chain Fv antibody that targets CCR5. Gene Ther. 2004;11:1627–1637. doi: 10.1038/sj.gt.3302329. [DOI] [PubMed] [Google Scholar]
- 192.Mukhtar M, Acheampong E, Khan MA, Bouhamdan M, Pomerantz RJ. Down-modulation of the CXCR4 co-receptor by intracellular expression of a single chain variable fragment (SFv) inhibits HIV-1 entry into primary human brain microvascular endothelial cells and post-mitotic neurons. Brain Res Mol Brain Res. 2005;135:48–57. doi: 10.1016/j.molbrainres.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 193.Swan CH, Buhler B, Steinberger P, Tschan MP, Barbas CF, 3rd, Torbett BE. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther. 2006;13:1480–1492. doi: 10.1038/sj.gt.3302801. [DOI] [PubMed] [Google Scholar]
- 194.Villani ME, Roggero P, Bitti O, Benvenuto E, Franconi R. Immunomodulation of cucumber mosaic virus infection by intrabodies selected in vitro from a stable single-framework phage display library. Plant Mol Biol. 2005;58:305–316. doi: 10.1007/s11103-005-4091-0. [DOI] [PubMed] [Google Scholar]
- 195.Scholthof K-BG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, et al. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12:938–954. doi: 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Matic S, Noris E, Contin R, Marian D, Thompson J. Engineering partial resistance to cucumber mosaic virus in tobacco using intrabodies specific for the viral polymerase. Phytochemistry. 2019;162:99–108. doi: 10.1016/j.phytochem.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 197.Ghannam A, Kumari S, Muyldermans S, Abbady AQ. Camelid nanobodies with high affinity for broad bean mottle virus: a possible promising tool to immunomodulate plant resistance against viruses. Plant Mol Biol. 2015;87:355–369. doi: 10.1007/s11103-015-0282-5. [DOI] [PubMed] [Google Scholar]
- 198.Hemmer C, Djennane S, Ackerer L, Hleibieh K, Marmonier A, Gersch S, et al. Nanobody-mediated resistance to Grapevine fanleaf virus in plants. Plant Biotechnol J. 2018;16:660–671. doi: 10.1111/pbi.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Vascotto F, Campagna M, Visintin M, Cattaneo A, Burrone OR. Effects of intrabodies specific for rotavirus NSP5 during the virus replicative cycle. J Gen Virol. 2004;85:3285–3290. doi: 10.1099/vir.0.80075-0. [DOI] [PubMed] [Google Scholar]
- 200.Desselberger U. Rotaviruses. Virus Res. 2014;190:75–96. doi: 10.1016/j.virusres.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 201.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ashour J, Schmidt FI, Hanke L, Cragnolini J, Cavallari M, Altenburg A, et al. Intracellular expression of camelid single-domain antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. J Virol. 2015;89:2792–2800. doi: 10.1128/JVI.02693-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tome-Amat J, Ramos I, Amanor F, Fernandez-Sesma A, Ashour J. Influenza a virus utilizes low-affinity, high-avidity interactions with the nuclear import machinery to ensure infection and immune evasion. J Virol. 2019;93:e01046-18. doi: 10.1128/JVI.01046-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mackenzie JS, Dimmock NJ. A preliminary study of physiological characteristics of temperature-sensitive mutants of influenza virus. J Gen Virol. 1973;19:51–63. doi: 10.1099/0022-1317-19-1-51. [DOI] [PubMed] [Google Scholar]
- 205.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci USA. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.De Baets S, Verhelst J, Van den Hoecke S, Smet A, Schotsaert M, Job ER, et al. A GFP expressing influenza A virus to report in vivo tropism and protection by a matrix protein 2 ectodomain-specific monoclonal antibody. PLoS One. 2015;10:e0121491. doi: 10.1371/journal.pone.0121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Beer B, Kurth R, Bukreyev A. Characteristics of Filoviridae: marburg and Ebola viruses. Naturwissenschaften. 1999;86:8–17. doi: 10.1007/s001140050562. [DOI] [PubMed] [Google Scholar]
- 208.Languon S, Quaye O. Filovirus disease outbreaks: a chronological overview. Virology (Auck). 2019;10:1178122X19849927. doi: 10.1177/1178122X19849927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Darling TL, Sherwood LJ, Hayhurst A. Intracellular crosslinking of filoviral nucleoproteins with xintrabodies restricts viral packaging. Front Immunol. 2017;8:1197. doi: 10.3389/fimmu.2017.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sundquist WI, Kräusslich H-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev. 2013;254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Justiz Vaillant AA, Gulick PG. HIV disease. Treasure Island: StatPearls Publishing; 2019. [Google Scholar]
- 213.Levin R, Mhashilkar AM, Dorfman T, Bukovsky A, Zani C, Bagley J, et al. Inhibition of early and late events of the HIV-1 replication cycle by cytoplasmic Fab intrabodies against the matrix protein, p17. Mol Med. 1997;3:96–110. [PMC free article] [PubMed] [Google Scholar]
- 214.McDougal JS, Kennedy MS, Sligh JM, Cort SP, Mawle A, Nicholson JK. Binding of HTLV-III/LAV to T4 + T cells by a complex of the 110 K viral protein and the T4 molecule. Science. 1986;231:382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- 215.Poznansky MC, Foxall R, Mhashilkar A, Coker R, Jones S, Ramstedt U, et al. Inhibition of human immunodeficiency virus replication and growth advantage of CD4+ T cells from HIV-infected individuals that express intracellular antibodies against HIV-1 gp120 or Tat. Hum Gene Ther. 1998;9:487–496. doi: 10.1089/hum.1998.9.4-487. [DOI] [PubMed] [Google Scholar]
- 216.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL. Cells transfected with a non-neutralizing antibody gene are resistant to HIV infection: targeting the endoplasmic reticulum and trans-Golgi network. J Immunol Baltim Md. 1950;1998(160):1489–1496. [PubMed] [Google Scholar]
- 218.Goncalves J, Silva F, Freitas-Vieira A, Santa-Marta M, Malho R, Yang X, et al. Functional neutralization of HIV-1 Vif protein by intracellular immunization inhibits reverse transcription and viral replication. J Biol Chem. 2002;277:32036–32045. doi: 10.1074/jbc.M201906200. [DOI] [PubMed] [Google Scholar]
- 219.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 220.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 221.Vercruysse T, Pardon E, Vanstreels E, Steyaert J, Daelemans D. An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 rev prevents viral production. J Biol Chem. 2010;285:21768–21780. doi: 10.1074/jbc.M110.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Matz J, Herate C, Bouchet J, Dusetti N, Gayet O, Baty D, et al. Selection of intracellular single-domain antibodies targeting the HIV-1 Vpr protein by cytoplasmic yeast two-hybrid system. PLoS One. 2014;9:e113729. doi: 10.1371/journal.pone.0113729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Fabryova H, Strebel K. Vpr and its cellular interaction partners: R we there yet? Cells. 2019;8:E1310. doi: 10.3390/cells8111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mhashilkar AM, Bagley J, Chen SY, Szilvay AM, Helland DG, Marasco WA. Inhibition of HIV-1 Tat-mediated LTR transactivation and HIV-1 infection by anti-Tat single chain intrabodies. EMBO J. 1995;14:1542–1551. doi: 10.1002/j.1460-2075.1995.tb07140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mhashilkar AM, Biswas DK, LaVecchio J, Pardee AB, Marasco WA. Inhibition of human immunodeficiency virus type 1 replication in vitro by a novel combination of anti-Tat single-chain intrabodies and NF-kappa B antagonists. J Virol. 1997;71:6486–6494. doi: 10.1128/jvi.71.9.6486-6494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Braun SE, Taube R, Zhu Q, Wong FE, Murakami A, Kamau E, et al. In vivo selection of CD4(+) T cells transduced with a gamma-retroviral vector expressing a single-chain intrabody targeting HIV-1 tat. Hum Gene Ther. 2012;23:917–931. doi: 10.1089/hum.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bao L, Hannon C, Cruz-Mignoni A, Ptchelkine D, Sun M-Y, Miller A, et al. Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein. Sci Rep. 2017;7:16869. doi: 10.1038/s41598-017-16742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen B. Molecular mechanism of HIV-1 entry. Trends Microbiol. 2019;27:878–891. doi: 10.1016/j.tim.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Helma J, Schmidthals K, Lux V, Nuske S, Scholz AM, Krausslich H-G, et al. Direct and dynamic detection of HIV-1 in living cells. PLoS One. 2012;7:e50026. doi: 10.1371/journal.pone.0050026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Casini A, Olivieri M, Vecchi L, Burrone OR, Cereseto A. Reduction of HIV-1 infectivity through endoplasmic reticulum-associated degradation-mediated Env depletion. J Virol. 2015;89:2966–2971. doi: 10.1128/JVI.02634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Richman DD, Little SJ, Smith DM, Wrin T, Petropoulos C, Wong JK. HIV evolution and escape. Trans Am Clin Climatol Assoc. 2004;115:289–303. [PMC free article] [PubMed] [Google Scholar]
- 232.Rice AP. The HIV-1 Tat protein: mechanism of action and target for HIV-1 cure strategies. Curr Pharm Des. 2017;23:4098–4102. doi: 10.2174/1381612823666170704130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Aguilar LK, Rooney CM, Heslop HE. Lymphoproliferative disorders involving Epstein–Barr virus after hemopoietic stem cell transplantation. Curr Opin Oncol. 1999;11:96–101. doi: 10.1097/00001622-199903000-00004. [DOI] [PubMed] [Google Scholar]
- 234.Lunn RM, Jahnke GD, Rabkin CS. Tumour virus epidemiology. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160266. doi: 10.1098/rstb.2016.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Piche A, Kasono K, Johanning F, Curiel TJ, Curiel DT. Phenotypic knock-out of the latent membrane protein 1 of Epstein-Barr virus by an intracellular single-chain antibody. Gene Ther. 1998;5:1171–1179. doi: 10.1038/sj.gt.3300706. [DOI] [PubMed] [Google Scholar]
- 236.Gennari F, Mehta S, Wang Y, St Clair Tallarico A, Palu G, Marasco WA. Direct phage to intrabody screening (DPIS): demonstration by isolation of cytosolic intrabodies against the TES1 site of Epstein Barr virus latent membrane protein 1 (LMP1) that block NF-kappaB transactivation. J Mol Biol. 2004;335:193–207. doi: 10.1016/j.jmb.2003.09.073. [DOI] [PubMed] [Google Scholar]
- 237.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 238.Corte-Real S, Collins C, da Silva FA, Simas JP, Barbas CF, III, Chang Y, et al. Intrabodies targeting the Kaposi sarcoma-associated herpesvirus latency antigen inhibit viral persistence in lymphoma cells. Blood. 2005;106:3797–3802. doi: 10.1182/blood-2005-04-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li H, Wang H, Nicholas J. Detection of direct binding of human herpesvirus 8-encoded interleukin-6 (vIL-6) to both gp130 and IL-6 receptor (IL-6R) and identification of amino acid residues of vIL-6 important for IL-6R-dependent and -independent signaling. J Virol. 2001;75:3325–3334. doi: 10.1128/JVI.75.7.3325-3334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Suthaus J, Adam N, Grotzinger J, Scheller J, Rose-John S. Viral interleukin-6: structure, pathophysiology and strategies of neutralization. Eur J Cell Biol. 2011;90:495–504. doi: 10.1016/j.ejcb.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 241.Kovaleva M, Bussmeyer I, Rabe B, Grotzinger J, Sudarman E, Eichler J, et al. Abrogation of viral interleukin-6 (vIL-6)-induced signaling by intracellular retention and neutralization of vIL-6 with an anti-vIL-6 single-chain antibody selected by phage display. J Virol. 2006;80:8510–8520. doi: 10.1128/JVI.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Harper DM, DeMars LR. HPV vaccines—a review of the first decade. Gynecol Oncol. 2017;146:196–204. doi: 10.1016/j.ygyno.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 243.Harden ME, Munger K. Human papillomavirus molecular biology. Mutat Res Rev Mutat Res. 2017;772:3–12. doi: 10.1016/j.mrrev.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Accardi L, Dona MG, Di Bonito P, Giorgi C. Intracellular anti-E7 human antibodies in single-chain format inhibit proliferation of HPV16-positive cervical carcinoma cells. Int J Cancer. 2005;116:564–570. doi: 10.1002/ijc.21052. [DOI] [PubMed] [Google Scholar]
- 247.Accardi L, Donà MG, Mileo AM, Paggi MG, Federico A, Torreri P, et al. Retinoblastoma-independent antiproliferative activity of novel intracellular antibodies against the E7 oncoprotein in HPV 16-positive cells. BMC Cancer. 2011;11:17. doi: 10.1186/1471-2407-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Accardi L, Paolini F, Mandarino A, Percario Z, Di Bonito P, Di Carlo V, et al. In vivo antitumor effect of an intracellular single-chain antibody fragment against the E7 oncoprotein of human papillomavirus 16. Int J Cancer. 2014;134:2742–2747. doi: 10.1002/ijc.28604. [DOI] [PubMed] [Google Scholar]
- 249.Griffin H, Elston R, Jackson D, Ansell K, Coleman M, Winter G, et al. Inhibition of papillomavirus protein function in cervical cancer cells by intrabody targeting. J Mol Biol. 2006;355:360–378. doi: 10.1016/j.jmb.2005.10.077. [DOI] [PubMed] [Google Scholar]
- 250.Amici C, Visintin M, Verachi F, Paolini F, Percario Z, Di Bonito P, et al. A novel intracellular antibody against the E6 oncoprotein impairs growth of human papillomavirus 16-positive tumor cells in mouse models. Oncotarget. 2016;7:15539–15553. doi: 10.18632/oncotarget.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Steels A, Vannevel L, Zwaenepoel O, Gettemans J. Nb-induced stabilisation of p53 in HPV-infected cells. Sci Rep. 2019;9:12680. doi: 10.1038/s41598-019-49061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Venkatakrishnan B, Zlotnick A. The structural biology of hepatitis B virus: form and function. Annu Rev Virol. 2016;3:429–451. doi: 10.1146/annurev-virology-110615-042238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Walayat S, Ahmed Z, Martin D, Puli S, Cashman M, Dhillon S. Recent advances in vaccination of non-responders to standard dose hepatitis B virus vaccine. World J Hepatol. 2015;7:2503–2509. doi: 10.4254/wjh.v7.i24.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Rey D, Krantz V, Partisani M, Schmitt MP, Meyer P, Libbrecht E, et al. Increasing the number of hepatitis B vaccine injections augments anti-HBs response rate in HIV-infected patients. Effects on HIV-1 viral load. Vaccine. 2000;18:1161–1165. doi: 10.1016/s0264-410x(99)00389-8. [DOI] [PubMed] [Google Scholar]
- 255.Catherine F-X, Piroth L. Hepatitis B virus vaccination in HIV-infected people: a review. Hum Vaccines Immunother. 2017;13:1–10. doi: 10.1080/21645515.2016.1277844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.MacLachlan JH, Cowie BC. Hepatitis B virus epidemiology. Cold Spring Harb Perspect Med. 2015;5:a021410. doi: 10.1101/cshperspect.a021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zampino R, Boemio A, Sagnelli C, Alessio L, Adinolfi LE, Sagnelli E, et al. Hepatitis B virus burden in developing countries. World J Gastroenterol. 2015;21:11941–11953. doi: 10.3748/wjg.v21.i42.11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 259.Serruys B, Van Houtte F, Verbrugghe P, Leroux-Roels G, Vanlandschoot P. Llama-derived single-domain intrabodies inhibit secretion of hepatitis B virions in mice. Hepatology. 2009;49:39–49. doi: 10.1002/hep.22609. [DOI] [PubMed] [Google Scholar]
- 260.Serruys B, Van Houtte F, Farhoudi-Moghadam A, Leroux-Roels G, Vanlandschoot P. Production, characterization and in vitro testing of HBcAg-specific VHH intrabodies. J Gen Virol. 2010;91:643–652. doi: 10.1099/vir.0.016063-0. [DOI] [PubMed] [Google Scholar]
- 261.Walsh R, Nuttall S, Revill P, Colledge D, Cabuang L, Soppe S, et al. Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology. 2011;411:132–141. doi: 10.1016/j.virol.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 262.WHO . Global hepatitis report, 2017. Geneva: WHO; 2017. [Google Scholar]
- 263.Pol S, Lagaye S. The remarkable history of the hepatitis C virus. Genes Immun. 2019;20:436–446. doi: 10.1038/s41435-019-0066-z. [DOI] [PubMed] [Google Scholar]
- 264.Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol. 2013;369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- 265.Heintges T, zu Putlitz J, Wands JR. Characterization and binding of intracellular antibody fragments to the hepatitis C virus core protein. Biochem Biophys Res Commun. 1999;263:410–418. doi: 10.1006/bbrc.1999.1350. [DOI] [PubMed] [Google Scholar]
- 266.Karthe J, Tessmann K, Li J, Machida R, Daleman M, Haussinger D, et al. Specific targeting of hepatitis C virus core protein by an intracellular single-chain antibody of human origin. Hepatology. 2008;48:702–712. doi: 10.1002/hep.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Suzuki R, Saito K, Matsuda M, Sato M, Kanegae Y, Shi G, et al. Single-domain intrabodies against hepatitis C virus core inhibit viral propagation and core-induced NFkappaB activation. J Gen Virol. 2016;97:887–892. doi: 10.1099/jgv.0.000423. [DOI] [PubMed] [Google Scholar]
- 268.Halfon P, Locarnini S. Hepatitis C virus resistance to protease inhibitors. J Hepatol. 2011;55:192–206. doi: 10.1016/j.jhep.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 269.Li DK, Chung RT. Overview of direct-acting antiviral drugs and drug resistance of hepatitis C virus. Methods Mol Biol. 2019;1911:3–32. doi: 10.1007/978-1-4939-8976-8_1. [DOI] [PubMed] [Google Scholar]
- 270.Gal-Tanamy M, Zemel R, Bachmatov L, Jangra RK, Shapira A, Villanueva RA, et al. Inhibition of protease-inhibitor-resistant hepatitis C virus replicons and infectious virus by intracellular intrabodies. Antiviral Res. 2010;88:95–106. doi: 10.1016/j.antiviral.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Alvarez RD, Barnes MN, Gomez-Navarro J, Wang M, Strong TV, Arafat W, et al. A cancer gene therapy approach utilizing an anti-erbB-2 single-chain antibody-encoding adenovirus (AD21): a phase I trial. Clin Cancer Res Off J Am Assoc Cancer Res. 2000;6:3081–3087. [PubMed] [Google Scholar]
- 272.Weidle UH, Maisel D, Brinkmann U, Tiefenthaler G. The translational potential for target validation and therapy using intracellular antibodies in oncology. Cancer Genom Proteom. 2013;10:239–250. [PubMed] [Google Scholar]
- 273.Buchfellner A, Yurlova L, Nuske S, Scholz AM, Bogner J, Ruf B, et al. A new nanobody-based biosensor to study endogenous PARP1 in vitro and in live human cells. PLoS One. 2016;11:e0151041. doi: 10.1371/journal.pone.0151041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Liu Y, Majumder S, McCall W, Sartor CI, Mohler JL, Gregory CW, et al. Inhibition of HER-2/neu kinase impairs androgen receptor recruitment to the androgen responsive enhancer. Cancer Res. 2005;65:3404–3409. doi: 10.1158/0008-5472.CAN-04-4292. [DOI] [PubMed] [Google Scholar]
- 275.Amundson SA, Myers TG, Fornace AJ. Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287–3299. doi: 10.1038/sj.onc.1202576. [DOI] [PubMed] [Google Scholar]
- 276.Liu T, Kuwana T, Zhang H, Vander Heiden MG, Lerner RA, Newmeyer DD. Phenotypic selection with an intrabody library reveals an anti-apoptotic function of PKM2 requiring Mitofusin-1. PLoS Biol. 2019;17:e2004413. doi: 10.1371/journal.pbio.2004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv. 2011;11:18–26. doi: 10.1124/mi.11.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Koo MY, Park J, Lim JM, Joo SY, Shin S-P, Shim HB, et al. Selective inhibition of the function of tyrosine-phosphorylated STAT3 with a phosphorylation site-specific intrabody. Proc Natl Acad Sci USA. 2014;111:6269–6274. doi: 10.1073/pnas.1316815111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Koistinen P, Heino J. The selective regulation of alpha Vbeta 1 integrin expression is based on the hierarchical formation of alpha V-containing heterodimers. J Biol Chem. 2002;277:24835–24841. doi: 10.1074/jbc.M203149200. [DOI] [PubMed] [Google Scholar]
- 281.Thammasit P, Sangboonruang S, Suwanpairoj S, Khamaikawin W, Intasai N, Kasinrerk W, et al. Intracellular acidosis promotes mitochondrial apoptosis pathway: role of EMMPRIN down-regulation via specific single-chain Fv intrabody. J Cancer. 2015;6:276–286. doi: 10.7150/jca.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Panich T, Tragoolpua K, Pata S, Tayapiwatana C, Intasai N. Downregulation of extracellular matrix metalloproteinase inducer by scFv-M6-1B9 intrabody suppresses cervical cancer invasion through inhibition of urokinase-type plasminogen activator. Cancer Biother Radiopharm. 2017;32:1–8. doi: 10.1089/cbr.2016.2126. [DOI] [PubMed] [Google Scholar]
- 283.Bertier L, Boucherie C, Zwaenepoel O, Vanloo B, Van Troys M, Van Audenhove I, et al. Inhibitory cortactin nanobodies delineate the role of NTA. FASEB J Off Publ Fed Am Soc Exp Biol. 2017;31:2460–2476. doi: 10.1096/fj.201600810RR. [DOI] [PubMed] [Google Scholar]
- 284.Bertier L, Hebbrecht T, Mettepenningen E, De Wit N, Zwaenepoel O, Verhelle A, et al. Nanobodies targeting cortactin proline rich, helical and actin binding regions downregulate invadopodium formation and matrix degradation in SCC-61 cancer cells. Biomed Pharmacother Biomed Pharmacother. 2018;102:230–241. doi: 10.1016/j.biopha.2018.03.064. [DOI] [PubMed] [Google Scholar]
- 285.Wheeler YY, Kute TE, Willingham MC, Chen S-Y, Sane DC. Intrabody-based strategies for inhibition of vascular endothelial growth factor receptor-2: effects on apoptosis, cell growth, and angiogenesis. FASEB J. 2003;17:1733–1735. doi: 10.1096/fj.02-0942fje. [DOI] [PubMed] [Google Scholar]
- 286.Boldicke T, Weber H, Mueller PP, Barleon B, Bernal M. Novel highly efficient intrabody mediates complete inhibition of cell surface expression of the human vascular endothelial growth factor receptor-2 (VEGFR-2/KDR) J Immunol Methods. 2005;300:146–159. doi: 10.1016/j.jim.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 287.Jendreyko N, Popkov M, Rader C, Barbas CF., 3rd Phenotypic knockout of VEGF-R2 and Tie-2 with an intradiabody reduces tumor growth and angiogenesis in vivo. Proc Natl Acad Sci USA. 2005;102:8293–8298. doi: 10.1073/pnas.0503168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Popkov M, Jendreyko N, McGavern DB, Rader C, Barbas CF., 3rd Targeting tumor angiogenesis with adenovirus-delivered anti-Tie-2 intrabody. Cancer Res. 2005;65:972–981. [PubMed] [Google Scholar]
- 289.Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 290.Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am J Hematol. 2018;93:442–459. doi: 10.1002/ajh.25011. [DOI] [PubMed] [Google Scholar]
- 291.Dixon AS, Constance JE, Tanaka T, Rabbitts TH, Lim CS. Changing the subcellular location of the oncoprotein Bcr-Abl using rationally designed capture motifs. Pharm Res. 2012;29:1098–1109. doi: 10.1007/s11095-011-0654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Mo SP, Coulson JM, Prior IA. RAS variant signalling. Biochem Soc Trans. 2018;46:1325–1332. doi: 10.1042/BST20180173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Biocca S, Pierandrei-Amaldi P, Cattaneo A. Intracellular expression of anti-p21ras single chain Fv fragments inhibits meiotic maturation of xenopus oocytes. Biochem Biophys Res Commun. 1993;197:422–427. doi: 10.1006/bbrc.1993.2496. [DOI] [PubMed] [Google Scholar]
- 294.Cardinale A, Lener M, Messina S, Cattaneo A, Biocca S. The mode of action of Y13-259 scFv fragment intracellularly expressed in mammalian cells. FEBS Lett. 1998;439:197–202. doi: 10.1016/s0014-5793(98)01369-6. [DOI] [PubMed] [Google Scholar]
- 295.Chambers JS, Brend T, Rabbitts TH. Cancer cell killing by target antigen engagement with engineered complementary intracellular antibody single domains fused to pro-caspase3. Sci Rep. 2019;9:8553. doi: 10.1038/s41598-019-44908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Quevedo CE, Cruz-Migoni A, Bery N, Miller A, Tanaka T, Petch D, et al. Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat Commun. 2018;9:3169. doi: 10.1038/s41467-018-05707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Mohty M, Gautier J, Malard F, Aljurf M, Bazarbachi A, Chabannon C, et al. CD19 chimeric antigen receptor-T cells in B-cell leukemia and lymphoma: current status and perspectives. Leukemia. 2019;33:2767–2778. doi: 10.1038/s41375-019-0615-5. [DOI] [PubMed] [Google Scholar]
- 298.Zhang C, Oberoi P, Oelsner S, Waldmann A, Lindner A, Tonn T, et al. chimeric antigen receptor-engineered NK-92 cells: an off-the-shelf cellular therapeutic for targeted elimination of cancer cells and induction of protective antitumor immunity. Front Immunol. 2017;8:533. doi: 10.3389/fimmu.2017.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 300.Zhang C, Helmsing S, Zagrebelsky M, Schirrmann T, Marschall ALJ, Schüngel M, et al. Suppression of p75 neurotrophin receptor surface expression with intrabodies influences Bcl-xL mRNA expression and neurite outgrowth in PC12 cells. PLoS One. 2012;7:e30684. doi: 10.1371/journal.pone.0030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ordonez A, Perez J, Tan L, Dickens JA, Motamedi-Shad N, Irving JA, et al. A single-chain variable fragment intrabody prevents intracellular polymerization of Z alpha1-antitrypsin while allowing its antiproteinase activity. FASEB J. 2015;29:2667–2678. doi: 10.1096/fj.14-267351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Bala G, Crauwels M, Blykers A, Remory I, Marschall ALJ, Dubel S, et al. Radiometal-labeled anti-VCAM-1 nanobodies as molecular tracers for atherosclerosis—impact of radiochemistry on pharmacokinetics. Biol Chem. 2019;400:323–332. doi: 10.1515/hsz-2018-0330. [DOI] [PubMed] [Google Scholar]
- 303.Godwin CD, Gale RP, Walter RB. Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia. 2017;31:1855–1868. doi: 10.1038/leu.2017.187. [DOI] [PubMed] [Google Scholar]
- 304.Connors JM, Jurczak W, Straus DJ, Ansell SM, Kim WS, Gallamini A, et al. Brentuximab vedotin with chemotherapy for stage III or IV Hodgkin’s lymphoma. N Engl J Med. 2018;378:331–344. doi: 10.1056/NEJMoa1708984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.von Minckwitz G, Huang C-S, Mano MS, Loibl S, Mamounas EP, Untch M, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 306.Kantarjian HM, DeAngelo DJ, Stelljes M, Liedtke M, Stock W, Gokbuget N, et al. Inotuzumab ozogamicin versus standard of care in relapsed or refractory acute lymphoblastic leukemia: final report and long-term survival follow-up from the randomized, phase 3 INO-VATE study. Cancer. 2019;125:2474–2487. doi: 10.1002/cncr.32116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Zehner M, Marschall AL, Bos E, Schloetel J-G, Kreer C, Fehrenschild D, et al. The translocon protein Sec61 mediates antigen transport from endosomes in the cytosol for cross-presentation to CD8(+) T cells. Immunity. 2015;42:850–863. doi: 10.1016/j.immuni.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 308.Reed SD, Li S. Electroporation advances in large animals. Curr Gene Ther. 2009;9:316–326. doi: 10.2174/156652309788921062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kvam E, Nannenga BL, Wang MS, Jia Z, Sierks MR, Messer A. Conformational targeting of fibrillar polyglutamine proteins in live cells escalates aggregation and cytotoxicity. PLoS One. 2009;4:e5727. doi: 10.1371/journal.pone.0005727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Butler DC, Snyder-Keller A, De Genst E, Messer A. Differential nuclear localization of complexes may underlie in vivo intrabody efficacy in Huntington’s disease. Protein Eng Des Sel PEDS. 2014;27:359–363. doi: 10.1093/protein/gzu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Martinelli C, Colombo E, Piccini D, Sironi C, Pelicci PG, de Marco A. An intrabody specific for the nucleophosmin carboxy-terminal mutant and fused to a nuclear localization sequence binds its antigen but fails to relocate it in the nucleus. Biotechnol Rep Amst Neth. 2014;3:27–33. doi: 10.1016/j.btre.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Simic G, Babic Leko M, Wray S, Harrington C, Delalle I, Jovanov-Milosevic N, et al. Tau protein hyperphosphorylation and aggregation in Alzheimer’s disease and other tauopathies, and possible neuroprotective strategies. Biomolecules. 2016;6:6. doi: 10.3390/biom6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ryan P, Xu M, Davey AK, Danon JJ, Mellick GD, Kassiou M, et al. O-GlcNAc modification protects against protein misfolding and aggregation in neurodegenerative disease. ACS Chem Neurosci. 2019;10:2209–2221. doi: 10.1021/acschemneuro.9b00143. [DOI] [PubMed] [Google Scholar]
- 314.Nizak C, Monier S, del Nery E, Moutel S, Goud B, Perez F. Recombinant antibodies to the small GTPase Rab6 as conformation sensors. Science. 2003;300:984–987. doi: 10.1126/science.1083911. [DOI] [PubMed] [Google Scholar]
- 315.Nizak C, Moutel S, Goud B, Perez F. Selection and application of recombinant antibodies as sensors of rab protein conformation. Methods Enzymol. 2005;403:135–153. doi: 10.1016/S0076-6879(05)03012-0. [DOI] [PubMed] [Google Scholar]
- 316.Galli V, Sebastian R, Moutel S, Ecard J, Perez F, Roux A. Uncoupling of dynamin polymerization and GTPase activity revealed by the conformation-specific nanobody dynab. eLife. 2017;6:e25197. doi: 10.7554/eLife.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Desplancq D, Freund G, Conic S, Sibler A-P, Didier P, Stoessel A, et al. Targeting the replisome with transduced monoclonal antibodies triggers lethal DNA replication stress in cancer cells. Exp Cell Res. 2016;342:145–158. doi: 10.1016/j.yexcr.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 318.Jendreyko N, Popkov M, Beerli RR, Chung J, McGavern DB, Rader C, et al. Intradiabodies, bispecific, tetravalent antibodies for the simultaneous functional knockout of two cell surface receptors. J Biol Chem. 2003;278:47812–47819. doi: 10.1074/jbc.M307002200. [DOI] [PubMed] [Google Scholar]
- 319.Cetin M, Evenson WE, Gross GG, Jalali-Yazdi F, Krieger D, Arnold D, et al. RasIns: genetically encoded intrabodies of activated Ras proteins. J Mol Biol. 2017;429:562–573. doi: 10.1016/j.jmb.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Brauchle M, Hansen S, Caussinus E, Lenard A, Ochoa-Espinosa A, Scholz O, et al. Protein interference applications in cellular and developmental biology using DARPins that recognize GFP and mCherry. Biol Open. 2014;3:1252–1261. doi: 10.1242/bio.201410041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Vernet E, Konrad A, Lundberg E, Nygren P-A, Graslund T. Affinity-based entrapment of the HER2 receptor in the endoplasmic reticulum using an affibody molecule. J Immunol Methods. 2008;338:1–6. doi: 10.1016/j.jim.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 322.Berglund DL, Starkey JR. Isolation of viable tumor cells following introduction of labelled antibody to an intracellular oncogene product using electroporation. J Immunol Methods. 1989;125:79–87. doi: 10.1016/0022-1759(89)90080-x. [DOI] [PubMed] [Google Scholar]
- 323.Kwee S, Nielsen HV, Celis JE. Electropermeabilization of human cultured cells grown in monolayers: incorporation of monoclonal antibodies. Bioelectrochem Bioenergy. 1990;23:65–80. [Google Scholar]
- 324.Wilson AK, Gorgas G, Claypool WD, de Lanerolle P. An increase or a decrease in myosin II phosphorylation inhibits macrophage motility. J Cell Biol. 1991;114:277–283. doi: 10.1083/jcb.114.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wilson AK, Horwitz J, De Lanerolle P. Evaluation of the electroinjection method for introducing proteins into living cells. Am J Physiol. 1991;260:C355–C363. doi: 10.1152/ajpcell.1991.260.2.C355. [DOI] [PubMed] [Google Scholar]
- 326.Berglund DL, Starkey JR. Introduction of antibody into viable cells using electroporation. Cytometry. 1991;12:64–67. doi: 10.1002/cyto.990120109. [DOI] [PubMed] [Google Scholar]
- 327.Kim D, Lee YJ, Rausch CM, Borrelli MJ. Electroporation of extraneous proteins into CHO cells: increased efficacy by utilizing centrifugal force and microsecond electrical pulses. Exp Cell Res. 1991;197:207–212. doi: 10.1016/0014-4827(91)90424-s. [DOI] [PubMed] [Google Scholar]
- 328.Dagher SF, Conrad SE, Werner EA, Patterson RJ. Phenotypic conversion of TK-deficient cells following electroporation of functional TK enzyme. Exp Cell Res. 1992;198:36–42. doi: 10.1016/0014-4827(92)90146-y. [DOI] [PubMed] [Google Scholar]
- 329.Maccarrone M, Veldink GA, Vliegenthart JF. Inhibition of lipoxygenase activity in lentil protoplasts by monoclonal antibodies introduced into the cells via electroporation. Eur J Biochem. 1992;205:995–1001. doi: 10.1111/j.1432-1033.1992.tb16866.x. [DOI] [PubMed] [Google Scholar]
- 330.Glogauer M, McCulloch CA. Introduction of large molecules into viable fibroblasts by electroporation: optimization of loading and identification of labeled cellular compartments. Exp Cell Res. 1992;200:227–234. doi: 10.1016/0014-4827(92)90168-8. [DOI] [PubMed] [Google Scholar]
- 331.Maccarrone M, Veldink GA, Finazzi Agro A, Vliegenthart JF. Lentil root protoplasts: a transient expression system suitable for coelectroporation of monoclonal antibodies and plasmid molecules. Biochim Biophys Acta. 1995;1243:136–142. doi: 10.1016/0304-4165(94)00124-g. [DOI] [PubMed] [Google Scholar]
- 332.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Wang J, Ladinsky MS, Howell KE. Molecules and vesicle coats involved in the budding of exocytotic vesicles from the trans-Golgi network. Cold Spring Harb Symp Quant Biol. 1995;60:139–146. doi: 10.1101/sqb.1995.060.01.017. [DOI] [PubMed] [Google Scholar]
- 334.Sung KL, Yang L, Whittemore DE, Shi Y, Jin G, Hsieh AH, et al. The differential adhesion forces of anterior cruciate and medial collateral ligament fibroblasts: effects of tropomodulin, talin, vinculin, and alpha-actinin. Proc Natl Acad Sci USA. 1996;93:9182–9187. doi: 10.1073/pnas.93.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Bright GR, Kuo NT, Chow D, Burden S, Dowe C, Przybylski RJ. Delivery of macromolecules into adherent cells via electroporation for use in fluorescence spectroscopic imaging and metabolic studies. Cytometry. 1996;24:226–233. doi: 10.1002/(SICI)1097-0320(19960701)24:3<226::AID-CYTO5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 336.Skouteris GG, Schroder CH. Cytosolic phospholipase A2 is activated by the hepatocyte growth factor receptor-kinase in Madin Darby canine kidney cells. J Cell Sci. 1997;110(Pt 14):1655–1663. doi: 10.1242/jcs.110.14.1655. [DOI] [PubMed] [Google Scholar]
- 337.Wang SM, Lo MC, Shang C, Kao SC, Tseng YZ. Role of M-line proteins in sarcomeric titin assembly during cardiac myofibrillogenesis. J Cell Biochem. 1998;71:82–95. doi: 10.1002/(sici)1097-4644(19981001)71:1<82::aid-jcb9>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 338.Wu JC, Chung TH, Tseng YZ, Wang SM. N-cadherin/catenin-based costameres in cultured chicken cardiomyocytes. J Cell Biochem. 1999;75:93–104. [PubMed] [Google Scholar]
- 339.Nakashima O, Terada Y, Hanada S, Yamamoto K, Kuwahara M, Sasaki S, et al. Activated STAT1 suppresses proliferation of cultured rat mesangial cells. Kidney Int. 2000;57:2249–2257. doi: 10.1046/j.1523-1755.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 340.Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- 341.Touyz RM, Wu XH, He G, Park JB, Chen X, Vacher J, et al. Role of c-Src in the regulation of vascular contraction and Ca2+ signaling by angiotensin II in human vascular smooth muscle cells. J Hypertens. 2001;19:441–449. doi: 10.1097/00004872-200103000-00012. [DOI] [PubMed] [Google Scholar]
- 342.Rui M, Chen Y, Zhang Y, Ma D. Transfer of anti-TFAR19 monoclonal antibody into HeLa cells by in situ electroporation can inhibit the apoptosis. Life Sci. 2002;71:1771–1778. doi: 10.1016/s0024-3205(02)01943-4. [DOI] [PubMed] [Google Scholar]
- 343.Lopez JJ, Salido GM, Pariente JA, Rosado JA. Interaction of STIM1 with endogenously expressed human canonical TRP1 upon depletion of intracellular Ca2+ stores. J Biol Chem. 2006;281:28254–28264. doi: 10.1074/jbc.M604272200. [DOI] [PubMed] [Google Scholar]
- 344.Jardin I, Lopez JJ, Salido GM, Rosado JA. Orai1 mediates the interaction between STIM1 and hTRPC1 and regulates the mode of activation of hTRPC1-forming Ca2+ channels. J Biol Chem. 2008;283:25296–25304. doi: 10.1074/jbc.M802904200. [DOI] [PubMed] [Google Scholar]
- 345.Todorova R. Estimation of methods of protein delivery into mammalian cells—a comparative study by electroporation and bioporter assay. Prikl Biokhim Mikrobiol. 2009;45:493–496. [PubMed] [Google Scholar]
- 346.Zbidi H, Jardin I, Woodard GE, Lopez JJ, Berna-Erro A, Salido GM, et al. STIM1 and STIM2 are located in the acidic Ca2+ stores and associates with Orai1 upon depletion of the acidic stores in human platelets. J Biol Chem. 2011;286:12257–12270. doi: 10.1074/jbc.M110.190694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Moeglin E, Desplancq D, Conic S, Oulad-Abdelghani M, Stoessel A, Chiper M, et al. Uniform widespread nuclear phosphorylation of histone H2AX is an indicator of lethal DNA replication stress. Cancers (Basel). 2019;11:E355. doi: 10.3390/cancers11030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Alex A, Piano V, Polley S, Stuiver M, Voss S, Ciossani G, et al. Electroporated recombinant proteins as tools for in vivo functional complementation, imaging and chemical biology. eLife. 2019;8:e48287. doi: 10.7554/eLife.48287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Scopa C, Marrocco F, Latina V, Ruggeri F, Corvaglia V, La Regina F, et al. Impaired adult neurogenesis is an early event in Alzheimer’s disease neurodegeneration, mediated by intracellular Aβ oligomers. Cell Death Differ. 2020;27(3):934–948. doi: 10.1038/s41418-019-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Wang C-E, Zhou H, McGuire JR, Cerullo V, Lee B, Li S-H, et al. Suppression of neuropil aggregates and neurological symptoms by an intracellular antibody implicates the cytoplasmic toxicity of mutant huntingtin. J Cell Biol. 2008;181:803–816. doi: 10.1083/jcb.200710158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Southwell AL, Bugg CW, Kaltenbach LS, Dunn D, Butland S, Weiss A, et al. Perturbation with intrabodies reveals that calpain cleavage is required for degradation of huntingtin exon 1. PLoS One. 2011;6:e16676. doi: 10.1371/journal.pone.0016676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Yamamoto M, Hayashi N, Takehara T, Ueda K, Mita E, Tatsumi T, et al. Intracellular single-chain antibody against hepatitis B virus core protein inhibits the replication of hepatitis B virus in cultured cells. Hepatology. 1999;30:300–307. doi: 10.1002/hep.510300105. [DOI] [PubMed] [Google Scholar]
- 353.Prabhu R, Khalap N, Burioni R, Clementi M, Garry RF, Dash S. Inhibition of hepatitis C virus nonstructural protein, helicase activity, and viral replication by a recombinant human antibody clone. Am J Pathol. 2004;165:1163–1173. doi: 10.1016/S0002-9440(10)63377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Flego M, Frau A, Accardi L, Mallano A, Ascione A, Gellini M, et al. Intracellular human antibody fragments recognizing the VP35 protein of Zaire Ebola filovirus inhibit the protein activity. BMC Biotechnol. 2019;19:64. doi: 10.1186/s12896-019-0554-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Owens RJ, Limn C, Roy P. Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. J Virol. 2004;78:6649–6656. doi: 10.1128/JVI.78.12.6649-6656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Li J, Zhang Q, Wang T, Li C, Liang M, Li D. Tracking hantavirus nucleocapsid protein using intracellular antibodies. Virol J. 2010;7:339. doi: 10.1186/1743-422X-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Hanke L, Schmidt FI, Knockenhauer KE, Morin B, Whelan SP, Schwartz TU, et al. Vesicular stomatitis virus N protein-specific single-domain antibody fragments inhibit replication. EMBO Rep. 2017;18:1027–1037. doi: 10.15252/embr.201643764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Liu Y, Sun L, Yu P, Li A, Li C, Tang Q, et al. Viral suppression function of intracellular antibody against C-terminal domain of rabies virus phosphoprotein. Acta Biochim Biophys Sin. 2015;47:815–823. doi: 10.1093/abbs/gmv060. [DOI] [PubMed] [Google Scholar]
- 359.Dekker S, Toussaint W, Panayotou G, de Wit T, Visser P, Grosveld F, et al. Intracellularly expressed single-domain antibody against p15 matrix protein prevents the production of porcine retroviruses. J Virol. 2003;77:12132–12139. doi: 10.1128/JVI.77.22.12132-12139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Wang L, Zhang L, Huang B, Li K, Hou G, Zhao Q, et al. A nanobody targeting viral nonstructural protein 9 inhibits porcine reproductive and respiratory syndrome virus replication. J Virol. 2019;93:e01888-18. doi: 10.1128/JVI.01888-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Blazek D, Celer V, Navratilova I, Skladal P. Generation and characterization of single-chain antibody fragments specific against transmembrane envelope glycoprotein gp46 of Maedi-Visna virus. J Virol Methods. 2004;115:83–92. doi: 10.1016/j.jviromet.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 362.Jiang W, Venugopal K, Gould EA. Intracellular interference of tick-borne flavivirus infection by using a single-chain antibody fragment delivered by recombinant Sindbis virus. J Virol. 1995;69:1044–1049. doi: 10.1128/jvi.69.2.1044-1049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Beerli RR, Wels W, Hynes NE. Intracellular expression of single chain antibodies reverts ErbB-2 transformation. J Biol Chem. 1994;269:23931–23936. [PubMed] [Google Scholar]
- 364.Hyland S, Beerli RR, Barbas CF, Hynes NE, Wels W. Generation and functional characterization of intracellular antibodies interacting with the kinase domain of human EGF receptor. Oncogene. 2003;22:1557–1567. doi: 10.1038/sj.onc.1206299. [DOI] [PubMed] [Google Scholar]
- 365.Figini M, Ferri R, Mezzanzanica D, Bagnoli M, Luison E, Miotti S, et al. Reversion of transformed phenotype in ovarian cancer cells by intracellular expression of anti folate receptor antibodies. Gene Ther. 2003;10:1018–1025. doi: 10.1038/sj.gt.3301962. [DOI] [PubMed] [Google Scholar]
- 366.Peng J-L, Wu S, Zhao X-P, Wang M, Li W-H, Shen X, et al. Downregulation of transferrin receptor surface expression by intracellular antibody. Biochem Biophys Res Commun. 2007;354:864–871. doi: 10.1016/j.bbrc.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 367.Newnham LE, Wright MJ, Holdsworth G, Kostarelos K, Robinson MK, Rabbitts TH, et al. Functional inhibition of beta-catenin-mediated Wnt signaling by intracellular VHH antibodies. mAbs. 2015;7:180–191. doi: 10.4161/19420862.2015.989023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Cruz-Migoni A, Canning P, Quevedo CE, Bataille CJR, Bery N, Miller A, et al. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc Natl Acad Sci USA. 2019;116:2545–2550. doi: 10.1073/pnas.1811360116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Redchuk TA, Karasev MM, Verkhusha PV, Donnelly SK, Hülsemann M, Virtanen J, et al. Optogenetic regulation of endogenous proteins. Nat Commun. 2020;11:605. doi: 10.1038/s41467-020-14460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Guillaume-Rousselet N, Jean D, Frade R. Cloning and characterization of anti-cathepsin L single chain variable fragment whose expression inhibits procathepsin L secretion in human melanoma cells. Biochem J. 2002;367:219–227. doi: 10.1042/BJ20020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Intasai N, Tragoolpua K, Pingmuang P, Khunkaewla P, Moonsom S, Kasinrerk W, et al. Potent inhibition of OKT3-induced T cell proliferation and suppression of CD147 cell surface expression in HeLa cells by scFv-M6-1B9. Immunobiology. 2008;214:410–421. doi: 10.1016/j.imbio.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 372.Delanote V, Vanloo B, Catillon M, Friederich E, Vandekerckhove J, Gettemans J. An alpaca single-domain antibody blocks filopodia formation by obstructing. FASEB J. 2010;24:105–118. doi: 10.1096/fj.09-134304. [DOI] [PubMed] [Google Scholar]
- 373.Van Impe K, Bethuyne J, Cool S, Impens F, Ruano-Gallego D, De Wever O, et al. A nanobody targeting the F-actin capping protein CapG restrains breast cancer metastasis. Breast Cancer Res. 2013;15:R116. doi: 10.1186/bcr3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Van Audenhove I, Boucherie C, Pieters L, Zwaenepoel O, Vanloo B, Martens E, et al. Stratifying fascin and cortactin function in invadopodium formation using inhibitory nanobodies and targeted subcellular delocalization. FASEB J. 2014;28:1805–1818. doi: 10.1096/fj.13-242537. [DOI] [PubMed] [Google Scholar]
- 375.Hebbrecht T, Van Audenhove I, Zwaenepoel O, Verhelle A, Gettemans J. VCA nanobodies target N-WASp to reduce invadopodium formation and functioning. PLoS One. 2017;12:e0185076. doi: 10.1371/journal.pone.0185076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Somplatzki S, Muhlenhoff M, Kroger A, Gerardy-Schahn R, Boldicke T. Intrabodies against the polysialyltransferases ST8SiaII and ST8SiaIV inhibit polysialylation of NCAM in rhabdomyosarcoma tumor cells. BMC Biotechnol. 2017;17:42. doi: 10.1186/s12896-017-0360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Beghein E, Devriese D, Van Hoey E, Gettemans J. Cortactin and fascin-1 regulate extracellular vesicle release by controlling endosomal trafficking or invadopodia formation and function. Sci Rep. 2018;8:15606. doi: 10.1038/s41598-018-33868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]