Figure 2. Cellular Hypoxia Adaptations.
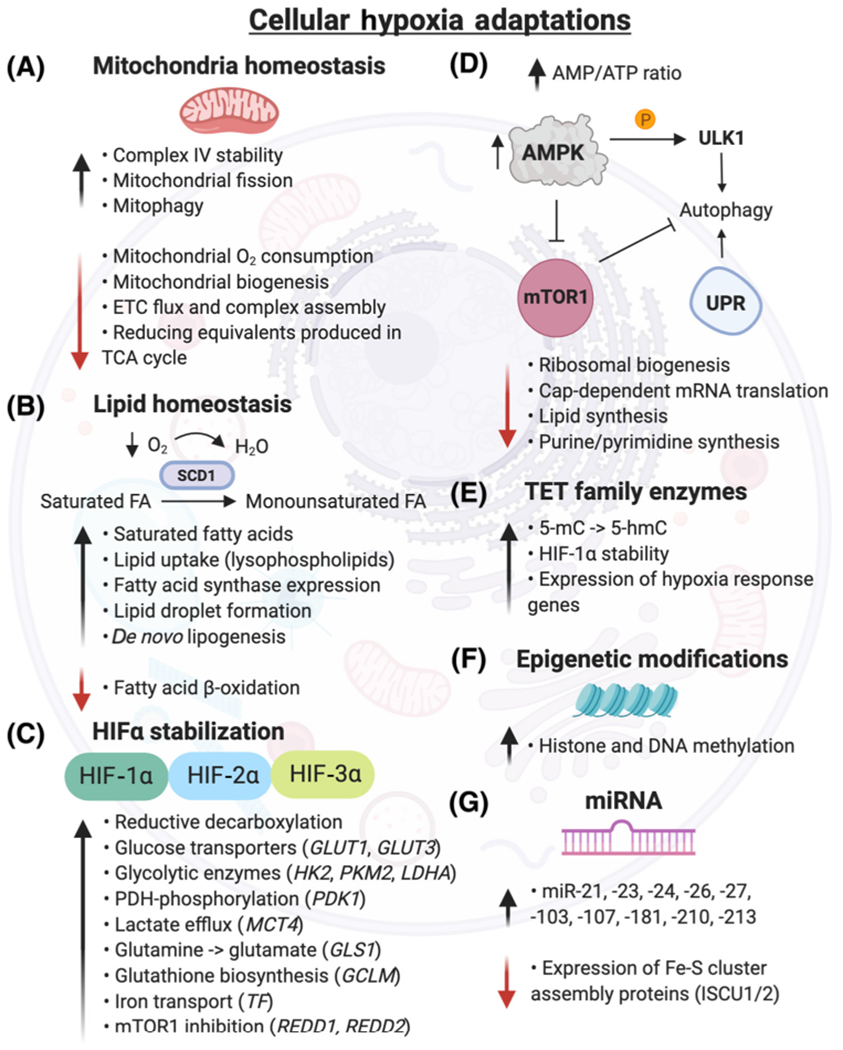
(A) Mitochondria adapt to hypoxia by increasing cytochrome c oxidase (complex IV) stability, which facilitates electron transport to O2. Mitochondria biogenesis, ETC flux, and respiration are downregulated, reducing O2 consumption and reactive oxygen species production [223]. Hypoxia induces mitochondrial fission by increasing the activity of the E3 ubiquitin ligase, SIAH2, which degrades AKAP121, subsequently increasing Drp1/Fis1 interaction [224]. (B) Under hypoxia, cells become deficient in unsaturated fatty acids by reduced O2-dependent desaturation of saturated fatty acids by stearoyl-CoA desaturases (e.g., SCD1). Hypoxic cells increase uptake of lysophospholipids and formation of lipid droplets, which serve as extra supply stores for lipids and help buffer against saturated lipid species [225]. Hypoxia downregulates fatty acid β-oxidation [226]. (C) Gene expression of several HIF targets are highlighted, though hundreds of genes are regulated by HIF stabilization [24]. (D) Hypoxia andthe subsequent energy deficiency (elevated AMP/ATP ratio) inhibit mTOR1 through AMP-activated protein kinase (AMPK). The downstream pathways regulated by mTOR1, including ribosomal and lipid synthesis, are downregulated in hypoxia, while apoptosis is increased. Unc-51-like kinase 1 (ULK1), an autophagy initiation serine/threonine protein kinase, is directly phosphorylated by AMPK in hypoxia, initiating a proautophagy signal [227]. Severe hypoxia (<0.01% O2) promotes the transcriptional response of essential autophagy genes and activates the UPR in a HIF-independent manner [228]. (E) Ten-eleven translocation methylcytosine dioxygenases (TET) catalyze the conversion of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC), increasing DNA methylation and gene expression of hypoxia responsive genes [8]. (F) Hypoxia induces changes in histone and DNA methylation via KDM and TET enzymes, respectively, which regulate chromatin and methylation homeostasis. (G) Numerous microRNAs are induced by hypoxia. miR-210 is increased in a HIF-1α-dependent manner and represses iron-sulfur cluster assembly proteins (ISCU1/2), resulting in decreased integrity of Fe-S cluster proteins [53]. Abbreviations: ETC, electron transport chain; FA, fatty acid; GCLM, glutamate-cysteine ligase modifier subunit; GLS1, glutaminase; GLUT, glucose transporter; HIF, hypoxia-inducible factor; HK2, hexokinase 2; KDM, lysine demethylase; LDHA, lactate dehydrogenase A; MCT4, monocarboxylate transporter 4; mTOR, mammalian target of rapamycin; PDK1, pyruvate dehydrogenase kinase; PKM2, pyruvate kinase muscle isozyme 2; REDD1/2, DNA damage response; TCA, tricarboxylic acid cycle; TF, transferrin; UPR, unfolded protein response.
