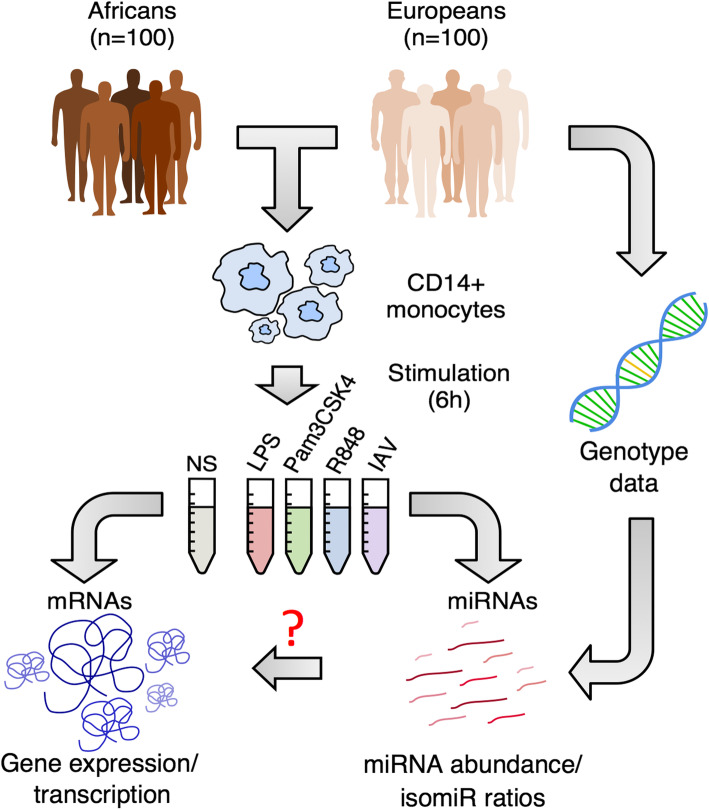
Fig. 1.
Population variation in miRNA response to immune activation. We stimulated monocytes from 200 healthy individuals using 3 TLR ligands as well as a strain of influenza A virus (IAV). For each individual, RNAs were extracted after 6 h of stimulation. We sequenced small RNAs, from both stimulated cells and non-stimulated cells (NS) (this study). The integration of miRNA sequencing data with genetic data, obtained through whole genome genotyping, whole-exome sequencing, and imputation [21], allowed us to assess the genetic bases of population differences in miRNA responses to stimulation, both quantitatively (miRNA abundance) and qualitatively (isomiR ratios). Furthermore, the availability of mRNA sequencing data from the same individuals and experimental conditions allows quantifying both total gene expression levels (exonic reads) and transcription rate (intronic reads derived from nascent mRNAs)