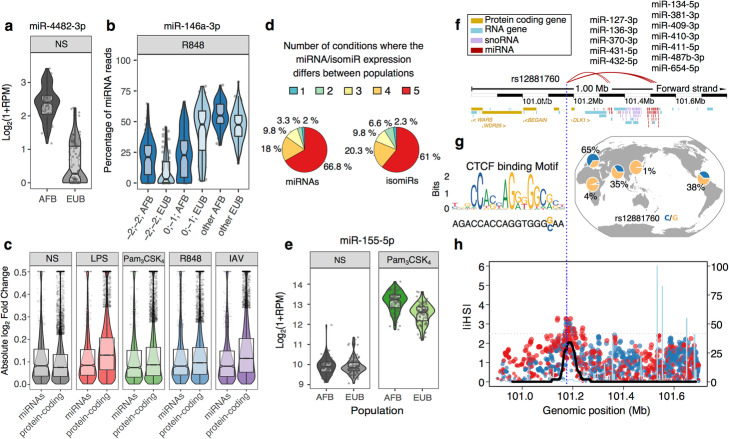
Fig. 5.
Population differences in miRNA expression. a Example of a miRNA (miR-4482-3p) differentially expressed between populations. Expression of miR-4482-3p is shown separately for African- (AFB) and European- (EUB) ancestry individuals. b IsomiRs of miR-146a-3p are differentially expressed between populations. For each population and isomiR, isomiR ratios are shown in the R848 condition where the difference is the strongest. All isomiRs with < 1 RPM on average, are pooled and annotated as other. c Amplitude of population differences in expression of miRNAs and protein-coding genes, at basal state and upon stimulation. d Sharing of pop-DE-miRs and pop-DE-isomiRs across conditions. For the 244 pop-DE-miRs and 188 pop-DE-isomiRs, number of conditions where we observed a difference between populations. e Expression of miR-155-5p is differential between African- and European-ancestry individuals, specifically in TLR-stimulated conditions. For simplicity, only Pam3CSK4 condition is shown. f–h Signatures of positive selection targeting the miR-QTL hotspot rs12881760. f Genomic context of the miR-QTL hotspot, displaying protein coding genes (yellow), RNA genes (cyan), snoRNAs (purple), and miRNAs (red) in a 1 Mb-window around the locus. Red lines link the miR-QTL to its target miRNAs, the name of which are indicated above. g Impact of the rs12881760 variant on a CTCF motif, and worldwide frequency of the motif-disrupting C allele. h Signatures of positive selection at the rs12881760 locus. |iHS| are displayed for all SNPs with MAF > 5% in Europeans, and dots are colored according to the sign of the iHS statistic (red, positive; blue, negative). The black line indicates the percentage of outliers (|iHS| > 2.5) on a sliding window of 100 consecutive SNPs with a MAF > 5% (right axis). Recombination rate is overlaid in light blue and normalized to the maximum recombination rate in the region (peak, 152 cM/Mb)