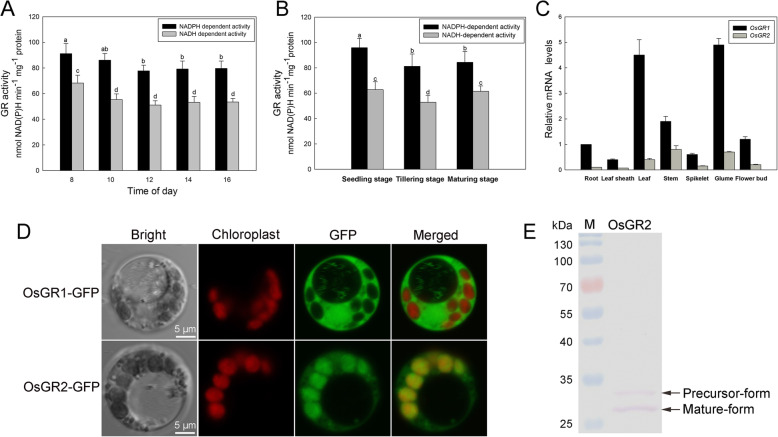
Fig. 1.
Expression patterns and subcellular localization of OsGR isoforms. a The diurnal changes of NAD(P)H-dependent GR activities in rice leaves. The second leaf from the top was detached from rice plants at the 5-leaf stage for determination. b The NAD(P)H-dependent GR activities at different developmental stages of rice plants. The second leaf from the top was detached for determination. c mRNA transcript abundances of OsGR1 and OsGR2 in different tissues were determined by qRT-PCR. Relative mRNA levels in various tissues were graphed based on the OsGR1 mRNA level in root as 1. Values are means ± SD of three replicates. Means denoted by the same letter did not significantly differ at P < 0.05 according to Duncan’s multiple range tests. d Subcellular localization of OsGR1 and OsGR2. The OsGR-GFP fusion constructs were transiently expressed in rice protoplasts, cells are imaged by a confocal microscope at 14–16 h after the transfection. The red signals represent chlorophyll autofluorescence. The result is representative of three independent experiments. Scale bar, 5 μm. e Proteins extracted from the OsGR2-GFP transformed protoplasts were analyzed by western blot using a monoclonal anti-GFP antibody. Precursor-form is the precursor protein of OsGR2-GFP synthesized in cytosol with an N-terminal CTP, Mature-form is the OsGR2-GFP located in choloroplast and the N-terminal CTP is cleaved off. The result is representative of three independent experiments