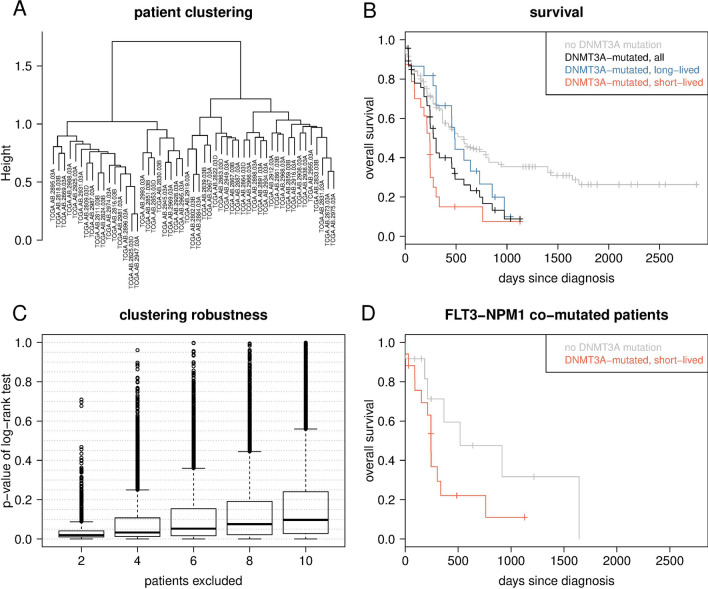
Figure 1.
Clustering of DNMT3A-mutated AML patients into two subgroups that differ in survival. (A) Hierarchical clustering of 51 DNMT3A-mutated AML patients; tip labels indicate TCGA identifiers (left subtree: short-lived, right subtree: long-lived). (B) Kaplan-Meier survival curves for the patients from (A) (black) and the two subgroups (short-lived: red, left subtree in A, survival data available for all 24 patients; long-lived: blue, right subtree in A, survival data available for 23 of 27 patients) as well as for 138 AML patients without a DNMT3A mutation (gray). Log-rank tests: P < 0.013 for red vs. blue, P < 0.0001 for red vs. grey, P = 0.345 for blue vs. grey, P = 0.004 for black vs. grey. (C) Robustness of clustering the DNMT3A-mutated patients into two subgroups that differ in survival, as assessed by randomly excluding patients and performing a hierarchical clustering and subsequent log-rank test on the data subset. Each boxplot shows the distributions of p-values of the log-rank tests for 10,000 data subsets. (D) Kaplan-Meier survival curves analyzing the impact of FLT3 and NPM1 co-mutations for all 17 affected patients of the short-lived subgroup (red) and all 12 affected patients of the 138 patients without a DNMT3A-mutation (grey). Log-rank test: P < 0.094.