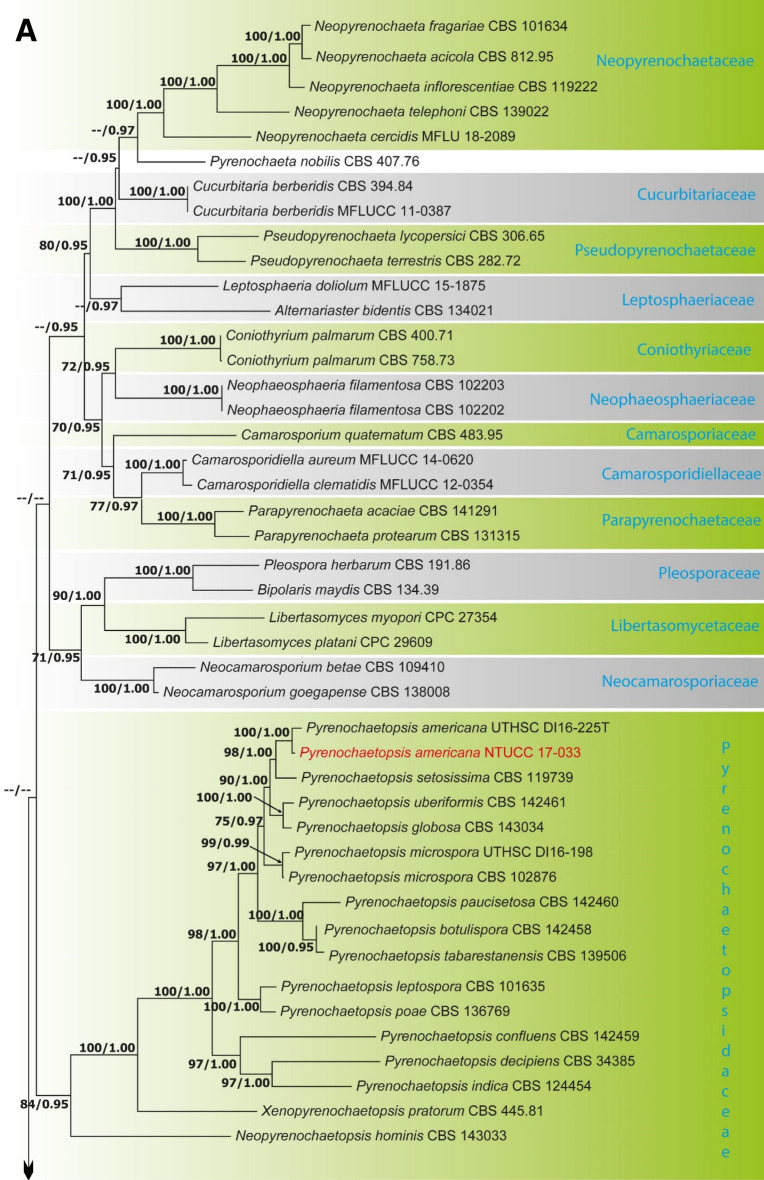
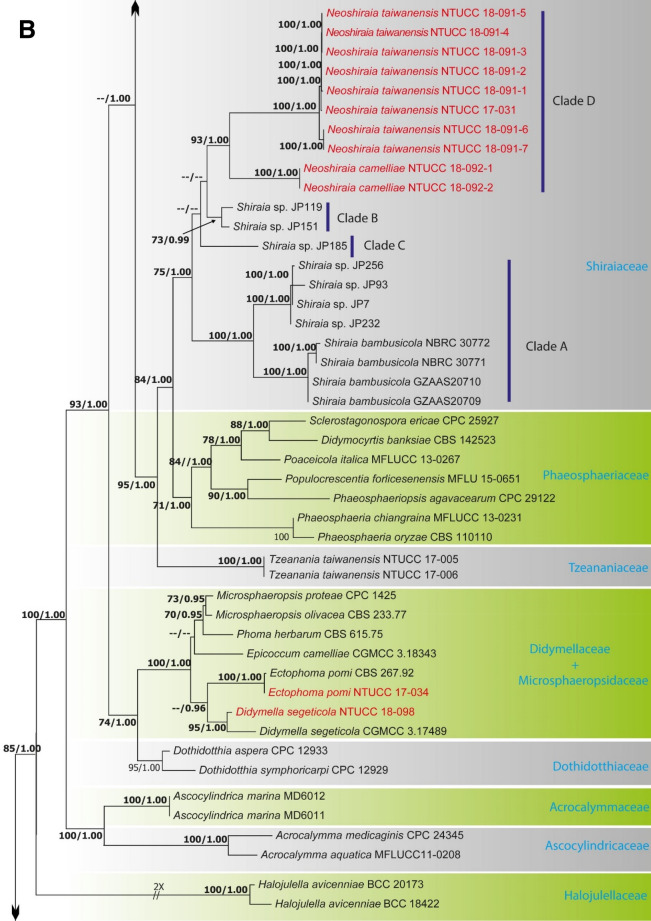
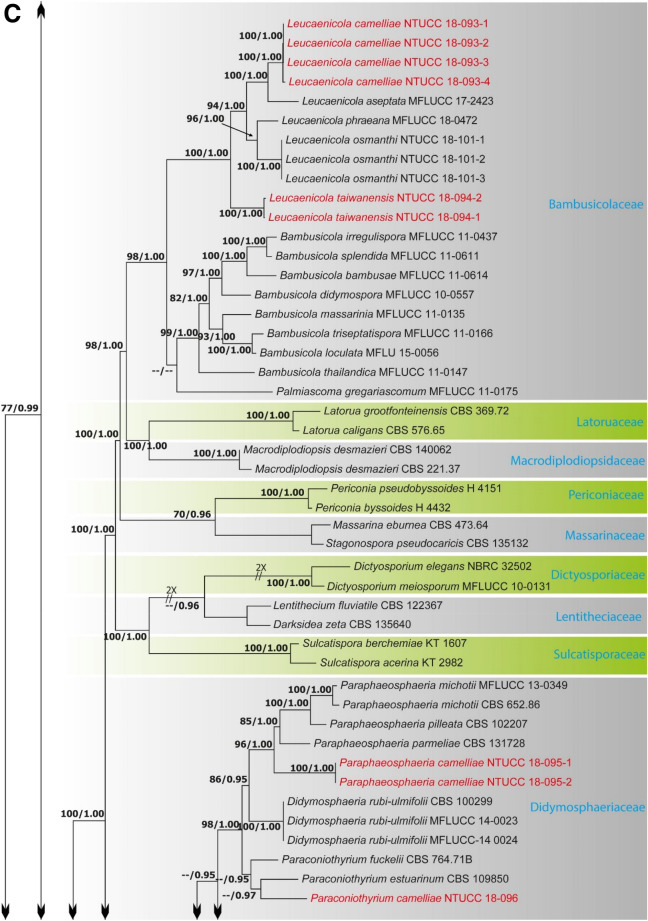
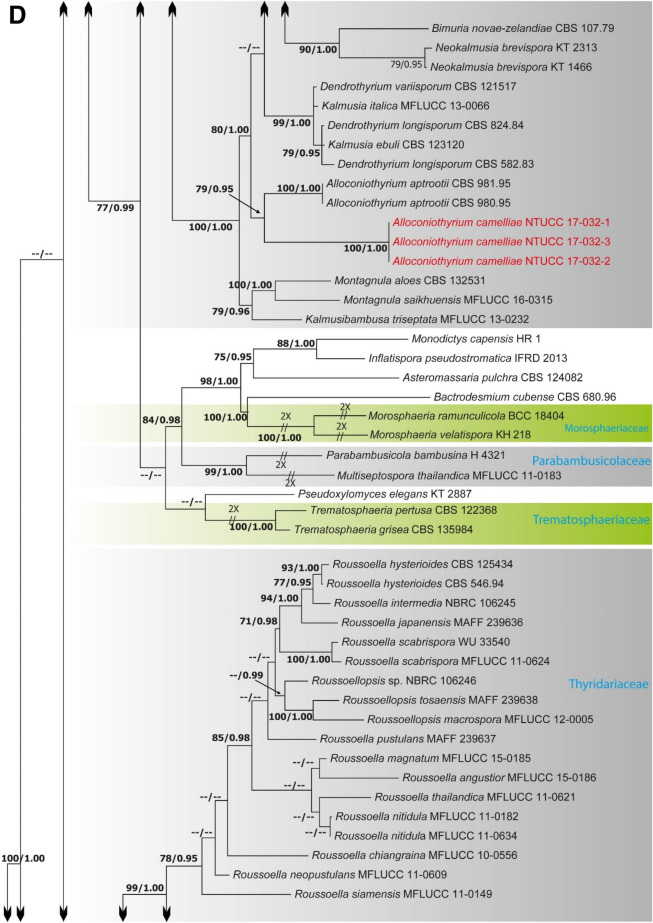
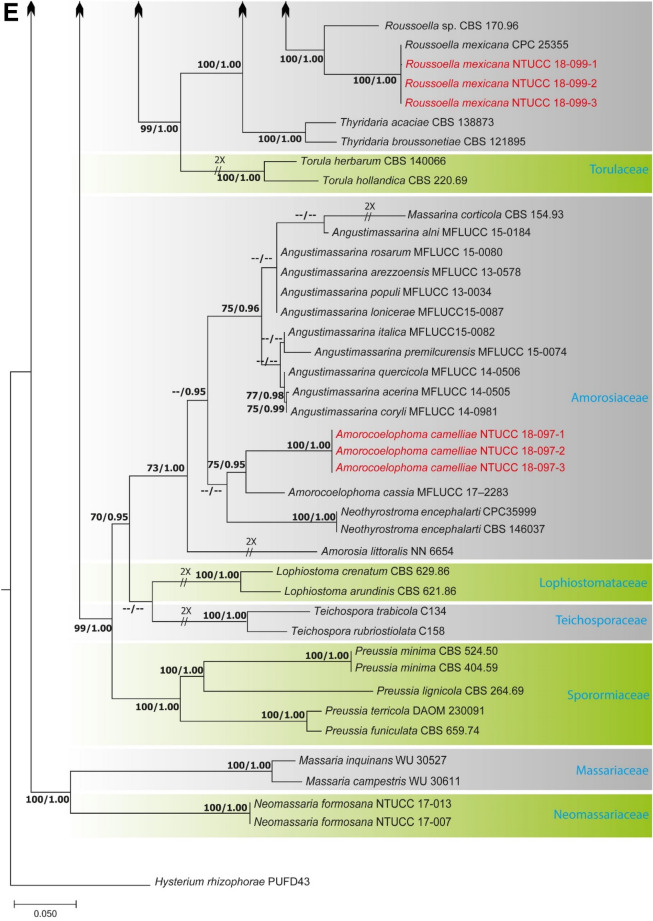
Figure 1.
(A) Part one of the phylogenetic tree based on the concatenated alignment of six molecular markers (ITS, LSU, SSU, rpb2, tef1 and tub2) evaluated using RAxML. The new isolates are shown in red. ML bootstrap values (MLBS) ≥ 70% and Bayesian posterior probabilities (PP) ≥ 0.95 are presented at the nodes. The scale bar indicates the number of estimated substitutions per site. Hysterium rhizophorae (PUFD43) was used as an outgroup for rooting the tree. (B) Part two, (C) Part three, (D) Part four, (E) Part five of the phylogenetic tree based on the concatenated alignment of six molecular markers (ITS, LSU, SSU, rpb2, tef1 and tub2) evaluated using RAxML.