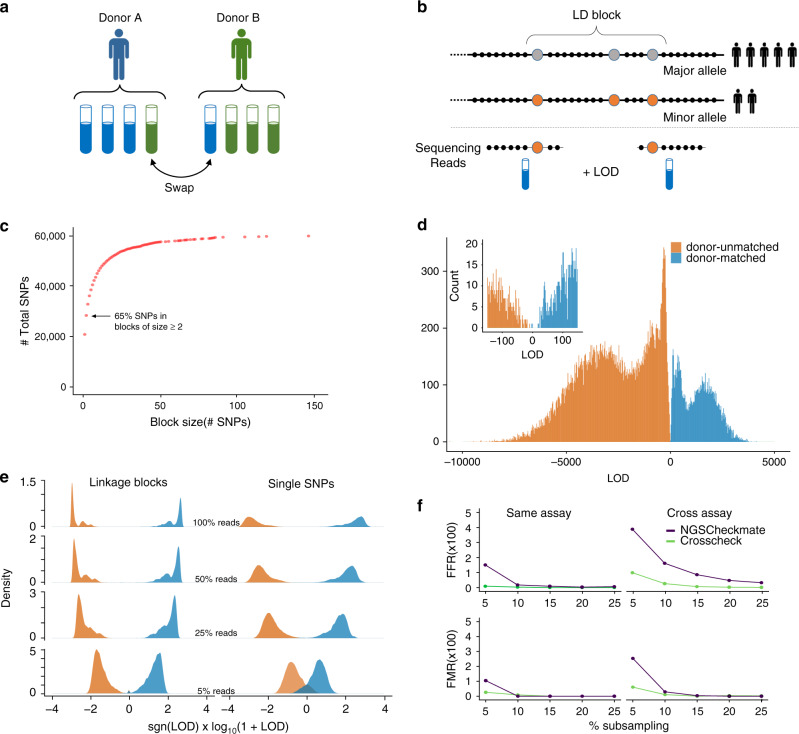
Fig. 1. Incorporating linkage information allows robust comparison of sequencing datasets.
a Sample swaps and misannotations, where a sample is incorrectly attributed to the wrong donor, are a high stakes issue for large consortium projects and clinical science. b Our method compares reads from two datasets across a genome-wide set of linkage disequilibrium (LD) blocks (haplotype map). The single-nucleotide polymorphisms (SNPs) in each block are highly correlated with each other and have low correlation with SNPs in other blocks. Reads overlapping any of the SNPs in a given block inform the relatedness of the datasets, even when reads from the two datasets do not overlap one another. c Haplotype maps contain many large LD blocks. LD blocks are created using common, ancestry independent SNPs from 1000 Genomes. Most SNPs lie within blocks of size >2, which boosts the chances of reads to be informative. d Distribution of LOD (log-odds ratio) scores for 34,336 donor-mismatched (red) and 9767 donor-matched pairs (green) of public ChIP-, RNA-, and DNase-seq datasets from the ENCODE project. e LD-based method can correctly determine sample relatedness even at low sequencing coverage. Pairwise comparisons of reference dataset pairs at different subsampling percentages using two equally sized SNP panels—one panel contained only independent single SNPs, while the other contained only LD blocks. Donor-mismatched dataset pairs are colored red while donor-matched dataset pairs are green. f Comparison of NGSC and Crosscheck’s classification of 34,336 donor-mismatched and 9767 donor-matched dataset pairs. Performance was measured in terms of the false flag rate (FFR), the fraction of donor-matched pairs incorrectly flagged as donor mismatches, and the false-match rate (FMR), the fraction of donor-mismatched pairs incorrectly identified as donor matches. Comparisons are classified as same-assay if the two datasets are from the same-assay type, and have the same target epitope in the case of ChIP-seq datasets. All other comparisons are classified as cross-assay. (Elements of (a and b) have been modified from a CDC publication (https://commons.wikimedia.org/wiki/File:Access_to_Health_Care-CDC_Vital_Signs-November_2010.pdf) which is under a CC BY-SA licence: https://creativecommons.org/licenses/by-sa/4.0/deed.en.)