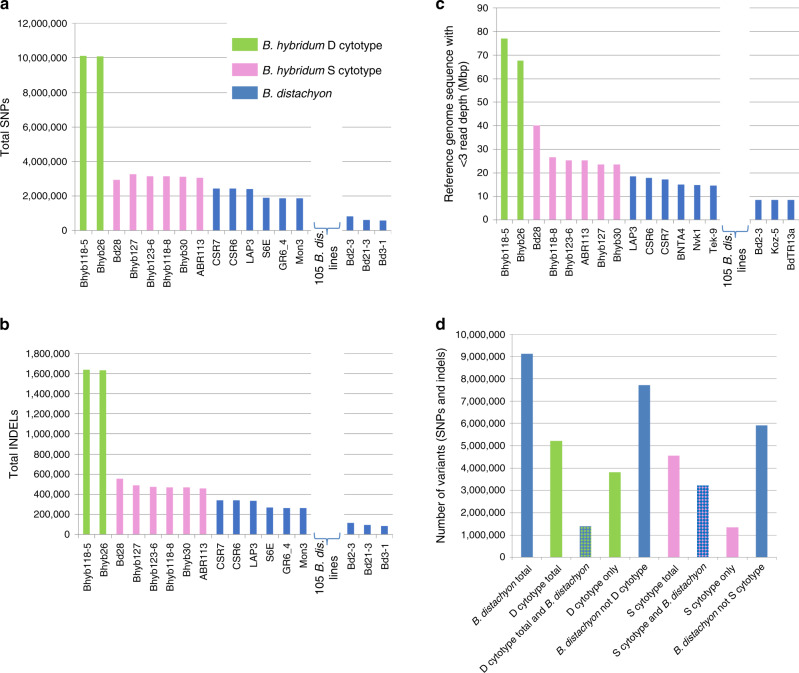
Fig. 8. Reference-based analysis.
Individual Illumina sequence reads, or simulated reads created by shredding a PacBio assembly (Bd28 accession), were mapped to the B. distachyon Bd21 reference genome to identify SNVs (a), small indels (b), and larger missing or highly divergent regions (c). In each graph, the lines have been ordered by descending number of polymorphisms, showing a trend from high levels in the B. hybridum D-plastotype lines, moderate levels in the B. hybridum-S-plastotype lines, and low levels in the B. distachyon lines. For clarity, 105 intermediate B. distachyon lines were omitted from the graphs. The B. hybridum D and S-plastotype lines both have significantly more SNVs, indels and deleted or highly divergent regions than B. distachyon (t tests, P < 10−20), whereas the B. hybridum D-plastotype lines had significantly more SNVs, indels (t test, P < 10−9) and deletions or highly divergent regions (t test, P < 10−4) than the S-plastotype lines. Note that the amount of deletion/highly divergent regions in Bd28 may be inflated by using a PacBio assembly that has more repetitive sequence assigned to the D subgenome than is possible by mapping Illumina reads. d The number of variants shared and unique to different groups. Green bars correspond to B. hybridum D plastotype, pink bars to B. hybridum-S plastotypes, and blue bars to B. distachyon accessions. Source data are provided as a Source Data file.