Fig 2. Template-based computational design.
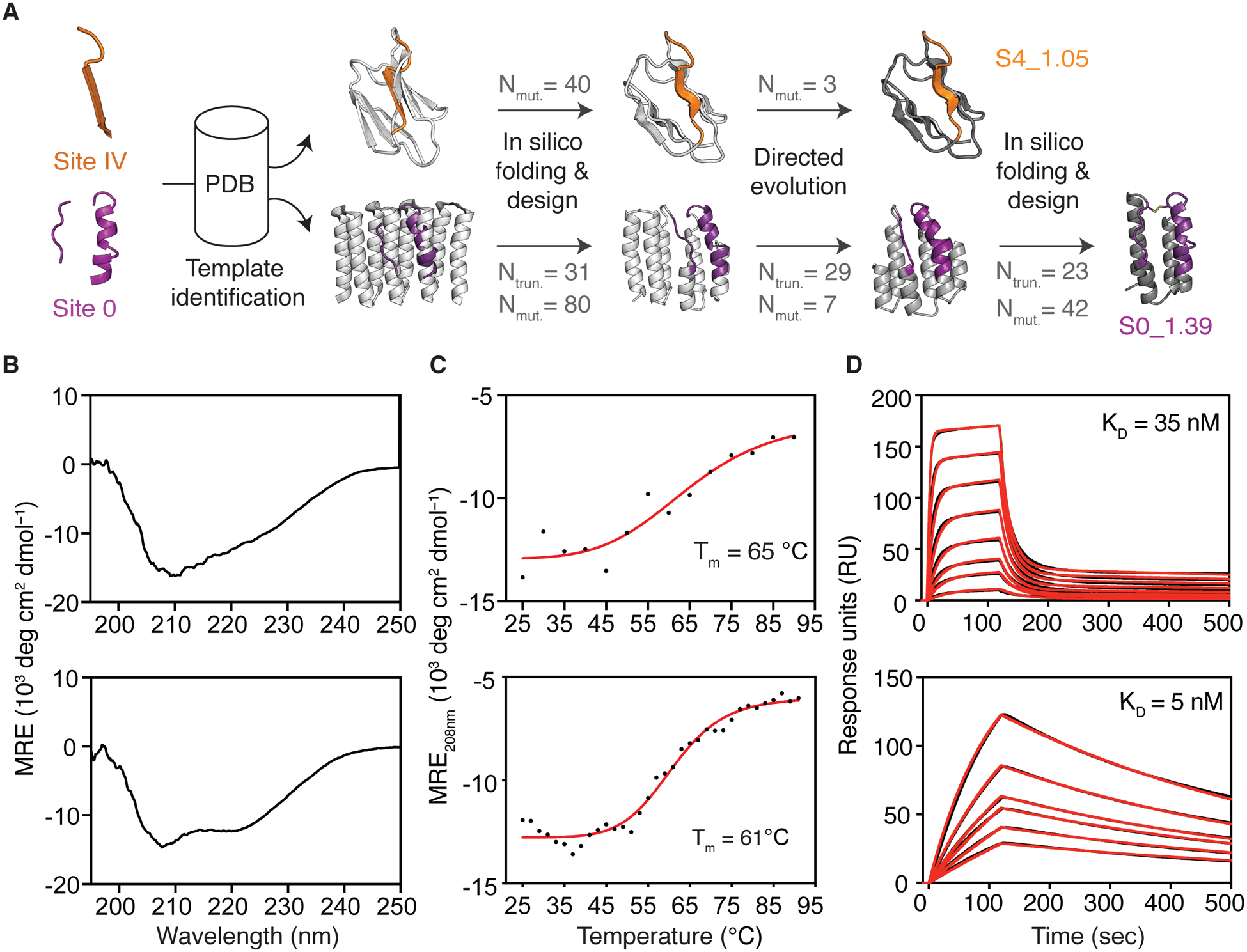
(A) Templates with structural similarity to sites IV and 0 were identified by native domain excision or loose structural matching, followed by in silico folding, design and directed evolution. Computational models of intermediates and final designs (S4_1.5 and S0_1.39) are shown, the number of mutations (Nmut) and truncated residues (Ntrun) are indicated for each step. (B) CD spectra measured at 20 °C of S4_1.5 (top) and S0_1.39 (bottom), are in agreement with the expected secondary structure content of the design models. (C) Thermal melting curves measured by CD in presence of reducing agent. (D) Binding affinity measured by SPR against target antibodies 101F (top) and D25 (bottom). Sensorgrams are shown in black and fits in red. CD: circular dichroism; Tm: melting temperature; SPR: surface plasmon resonance.
