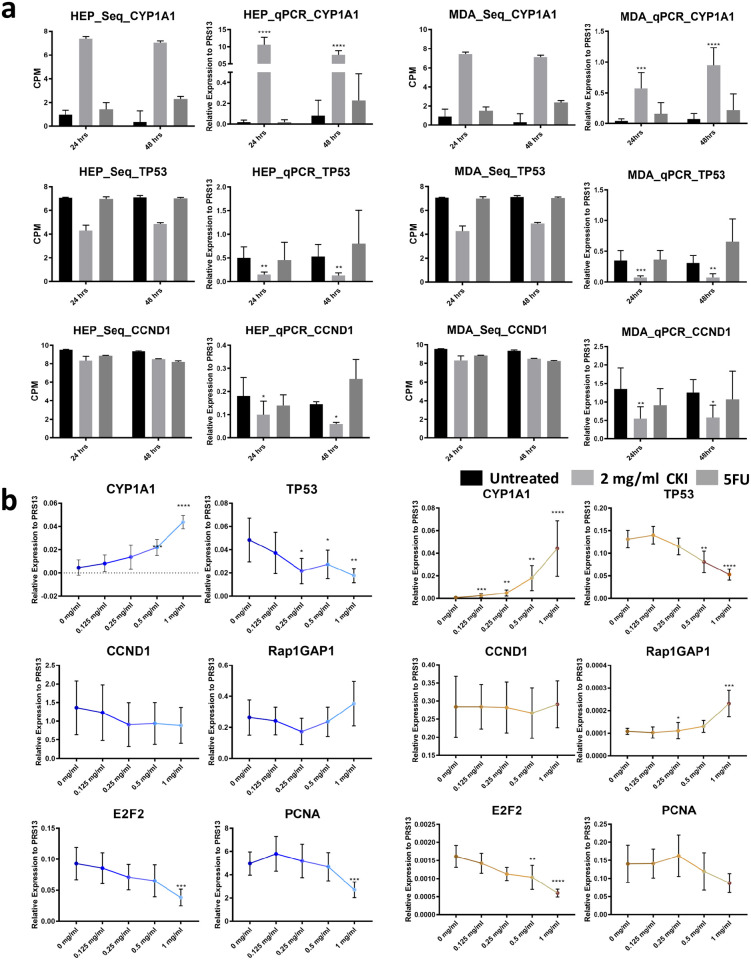
Fig 3. Validation of gene expression and effects of low dose CKI using RT-qPCR.
a: Comparison of DE genes between RNA-seq results (left) and RT-qPCR validation (right) for each cell line at 2 time-points. Three DE genes (CYP1A1, TP53 and CCND1) were chosen for validation. Gene expression was generally consistent between transcriptome data and qPCR data. b: Dose response of CKI using a subset of genes with conserved expression in Hep G2 (left), and MDA-MB-231 (right) from 0 mg/ml to 1 mg/ml of total alkaloids. Six genes (CYP1A1, TP53, CCND1, Rap2GAP1, E2F2 and PCNA) were selected based on their relevance to important pathways perturbed by CKI. RT-qPCR results are presented as expression relative to RPS13. Data are represented as mean ±95%CI (n>3). A t-test was used to compare CKI doses with “untreated” (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).