Figure 2.
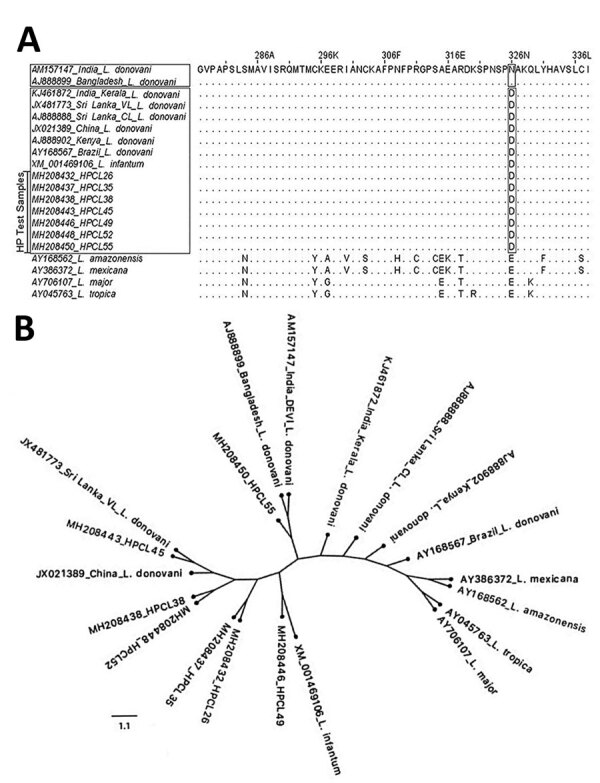
6PGDH-based molecular analysis of clinical isolates from cutaneous leishmaniasis (CL) patients, Himachal Pradesh, India, 2014–2018. A) Sequence alignment of partial 6PGDH amino acid of CL isolates exhibit replacement of asparagine (N) with aspartic acid (D) at position 326 analogous to visceral leishmaniasis–causing and CL-causing isolates from Sri Lanka. B) Phylogenetic tree for 6PGDH sequences from CL test isolates (designated as HPCL, numbered in order of their collection) and standard Leishmania strains. Tree constructed by using maximum-likelihood method with 5,000 bootstraps in the dnaml program of PHYLIP package (http://evolution.genetics.washington.edu/phylip/doc/main.html). GenBank accession numbers are indicated. Scale bar indicates the amino acid substitution per site. 6PGDH, 6-phosphogluconate dehydrogenase gene; HP, Himachal Pradesh.
