Figure 2.
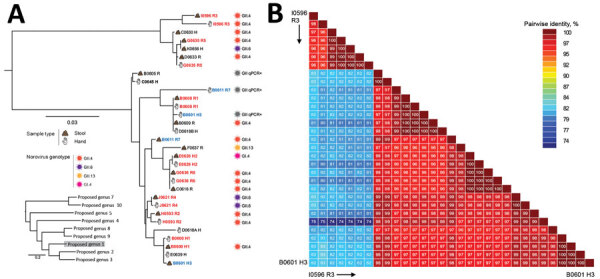
Phylogenetic relationships and pairwise sequence comparison of crAssphage strains from hand rinse samples collected during norovirus outbreaks at long-term care facilities. A) Phylogeny of crAssphage strains. Strain identification includes facility (A–J), strain number, and human source (H, healthcare worker; R, resident) for each isolate. Sample source and genotypes are indicated. Red strain names indicate that both hand and stool sample are genetically related, blue strain names that paired hand and stool samples are genetically distinct. Black strain names indicate hand or stool sample pairs that tested negative for crAssphage. Inset shows position of long-term care strains among reference strains; scale bar indicates number of nucleotide changes between sequences. (B) Color-coded pairwise identity matrix for crAssphage strains. Each cell includes the percentage identity among 2 sequences (horizontally to the left and vertically at the bottom). Key indicates pairwise identity percentages
