Figure 6.
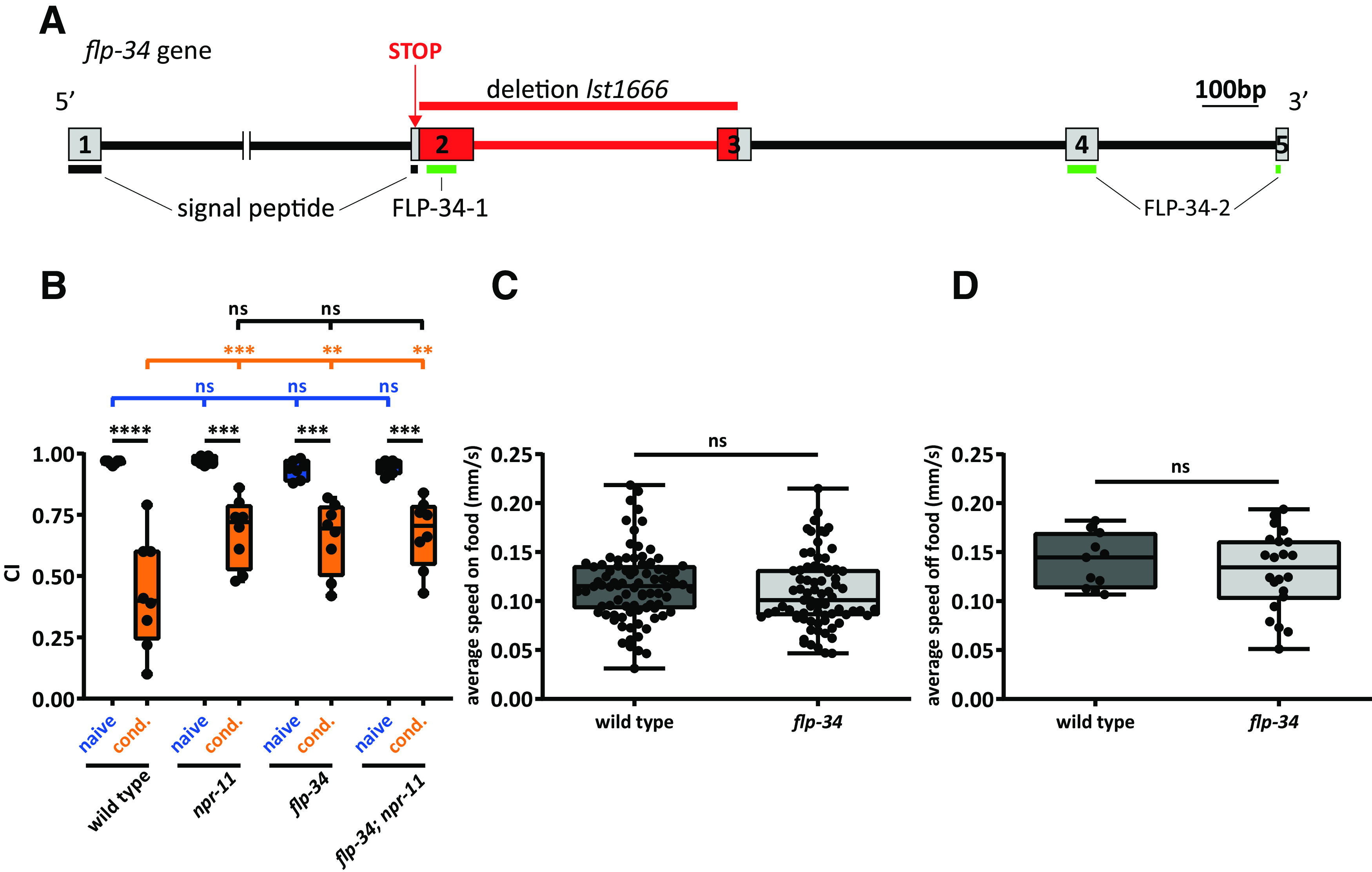
flp-34 mutants phenocopy the defect of npr-11 mutants in diacetyl learning. A, CRISPR/Cas9-induced deletion in a flp-34 loss-of-function mutant. Boxes represent exons encoding FLP-34, numbered 1-5. Lines between boxes indicate introns. Part of the intron between exons 1 and 2 is cropped. Red represents the CRISPR/Cas9 deletion (lst1666). A stop codon introduced in exon 2 interrupts translation of the signal peptide region, preventing the synthesis of downstream sequences containing both mature peptides. Horizontal lines indicate signal peptide (black) and mature peptide sequences (green). Additional information about the CRISPR/Cas9-edited deletion can be found in Extended Data Figure 6-1. B, flp-34 mutants are defective in diacetyl learning. The CI of flp-34 mutants after conditioning is similar to that of conditioned npr-11 mutant animals and significantly different from WT. An npr-11;flp-34 double mutant does not display additive learning defects. Two-way ANOVA revealed a significant effect of genotypes (F(3,52) = 3.262, p = 0.0286), of behavioral treatments (F(1,52) = 116.1, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(3,52) = 4.048, p = 0.0116). Data were analyzed by Sidak's post hoc test (n ≥ 7). C, D, flp-34 mutants display normal locomotion speed on and off food. Activity is recorded for 10 min on OP50-seeded (C) or unseeded (D) NGM plates. The average speed of mutants is not significantly different from that of WT in both conditions. Data were analyzed by unpaired t test (C, w ≥ 75; D, w ≥ 11). B–D, Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) indicates the median. Whiskers represent the minimum and maximum values. Black dots represent individual CIs or worms. **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
