Figure 2. Some examples of lncRNA activity as a regulator of gene expression.
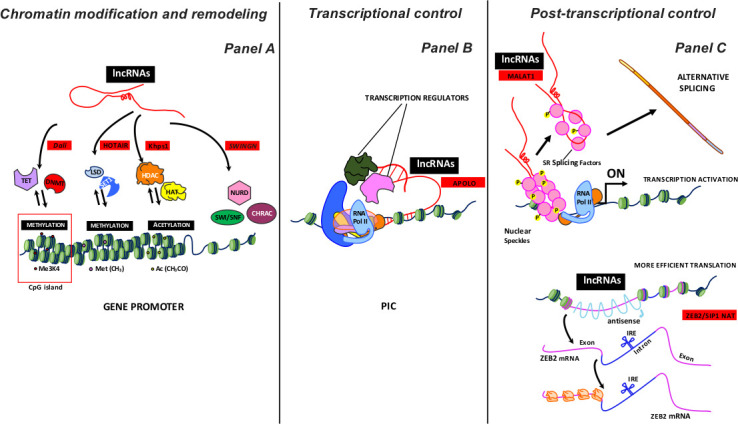
LncRNAs, working on proximal loci or at a great distance, can inhibit or activate the expression gene at various levels: i) chromatin modification and remodeling; ii) transcriptional control, and iii) post-transcriptional control. Panel A summarizes the different chromatin remodeling mechanisms. From left to right: nucleotide covalent modifiers (methylation) at CpG island (TET, DNMT enzymes); histone covalent modifiers, which regulate amino acid methylation (LSD, SET1 enzymes) and acetylation (HADC and HAT enzymes); ATP-dependent chromatin remodeling complexes (SWI/SNF, NURD, CHRAC). The lncRNAs acting with different enzymatic complexes are depicted with red squares. Panel B shows lncRNAs involved in the early transcription stages. LncRNAs acquiring a specific R-loops structure interact with PIC (Preinitiation Complex) to modulate transcription by recruiting regulator factors. The lncRNA acting with different transcription regulators is depicted with a red square. Panel C shows some post-transcription activities of lncRNAs. Top: MALAT1 localizes to interchromatin granule clusters (nuclear speckles) and regulates alternative splicing by modulating the distribution and the levels of active SR splicing factors. Bottom: ZEB2/SIP1 NAT binds and masks specific splicing sites, causing intron retention. The retained intron contains an IRES site (internal ribosome entry site) that induces a more efficient ZEB2 protein translation. The lncRNAs acting with different enzymatic complexes are depicted with red squares.
