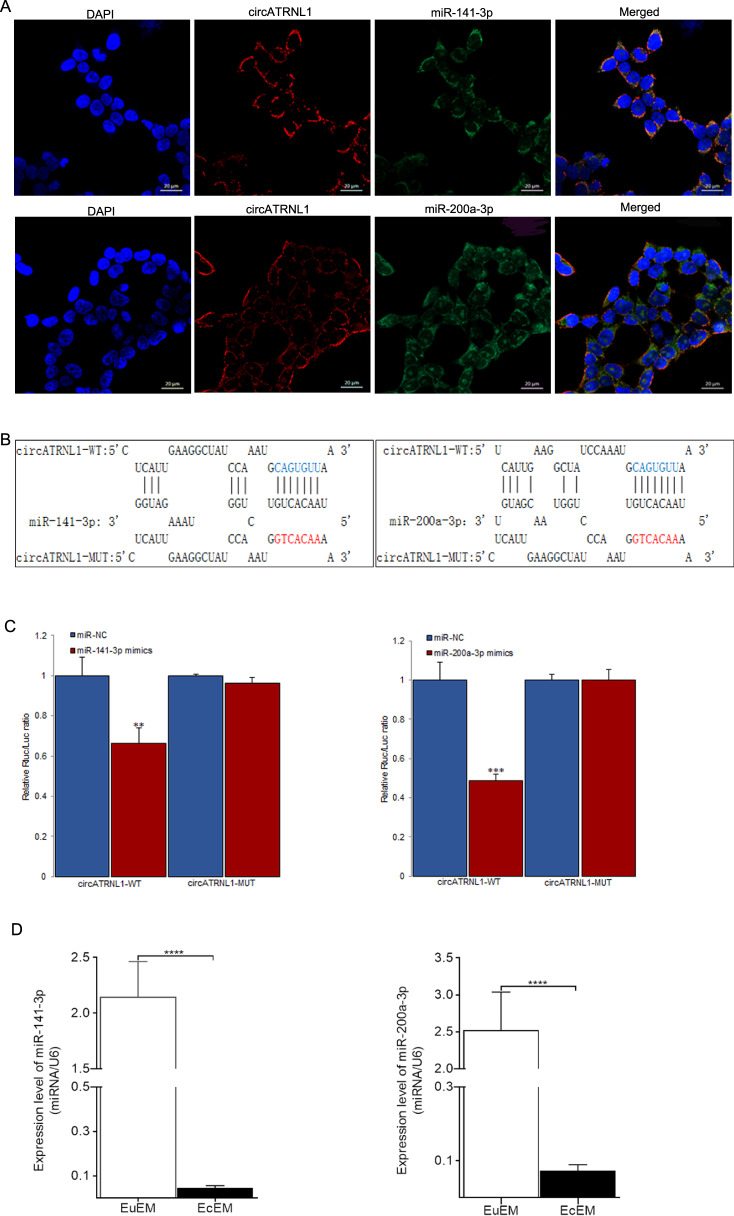
Fig. 2. miR-141-3p and miR-200a-3p are targets of circATRNL1.
a FISH assay was used to determine the location of circATRNL1, miR-141-3p, and miR-200a-3p in Ishikawa cells (blue, DAPI nuclear staining; red, circATRNL1; green, miR-141-3p and miR-200a-3p). Scale bars represent 20 μm. b The binding sites between wild-type circATRNL1 or mutated circATRNL1 and miR-141-3p/miR-200a-3p were predicted by using bioinformatics analysis. c Dual-luciferase reporter assays were carried out in HEK-293T cell to confirm the combination between circATRNL1 and miR-141-3p mimics or miR-200a-3p mimics. d qRT-PCR analysis was carried to determine the expression pattern of miR-141-3p and miR-200a-3p in ovary endometriosis tissues. ****P < 0.0001 represents statistical difference.