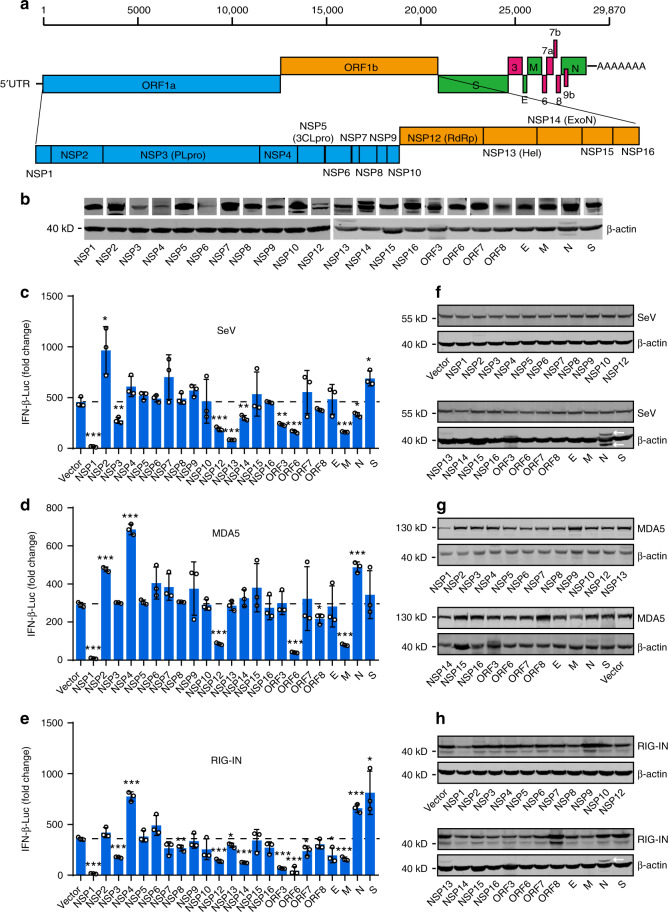
Fig. 2. Identification of viral proteins perturbing IFN-β production.
a Schematic diagrams of the SARS-CoV-2 genome. The genome includes 5′UTR-ORF1a-ORF1b-S-ORF3-E-M-ORF6-ORF7 (7a and 7b)-ORF8-N-3′UTR in order. Fifteen nonstructural proteins, four structural proteins, and four accessory proteins were delineated. b Protein expressions of SARS-CoV-2 genes. HEK293T cells were transfected with 500 ng plasmid in 24-well plates. Protein expressions were detected by Western blot. β-actin was used as a loading control. c Effect of SARS-CoV-2 proteins on SeV-induced IFN-β promoter activation. HEK293T cells were transfected with an IFN-β reporter plasmid, along with a control plasmid or with plasmids expressing the indicated SARS-CoV-2 proteins. At 24 h post-transfection, cells were infected with SeV for 12 h, and luciferase activity was measured. d, e Effects of SARS-CoV-2 proteins on RIG-IN and MDA5-induced IFN-β promoter activation. HEK293T cells were transfected with IFN-β promoter plasmid, along with a control plasmid or with plasmids expressing the indicated SARS-CoV-2 proteins, together with a plasmid expressing RIG-IN d or MDA5 e. At 24 h post-transfection, cells were harvested and luciferase activity was measured. f–h Expressions levels of SARS-CoV-2 protein, RIG-IN, and MDA5. Lysates of cells from c–e were subjected to Western blot analysis. Arrows indicate remnants of blots for SARS-CoV-2 proteins. All experiments were done at least twice, and one representative is shown. Error bars indicate SD of technical triplicates. *P < 0.05, **P < 0.01, and ***P < 0.001, two-tailed Student’s t-test. Source data are provided as a Source Data file.