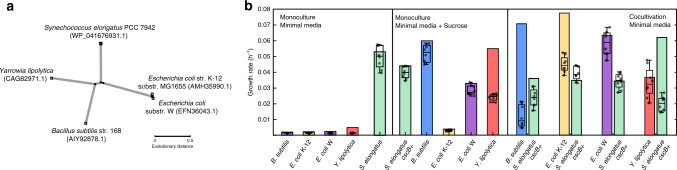
Fig. 1. Community members and growth performance.
a Neighbor-joining tree based of almost full-length 16S rRNA gene sequences, showing phylogenetic relationships between S. elongatus PCC7942 and the heterotrophic species used here. GenBank accession numbers are given in parentheses. b The bar plot depicts growth rates predicted using CM-models. Boxplots show experimental validation using at least six experimental replicates, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers. The first panel shows growth in monoculture using minimal media without an organic carbon source. The second panel shows growth rates during monoculture, while using minimal media with additional sucrose. The third panel shows results obtained when combining S. elongatus cscB+ with all heterotrophs in minimal media. Source data underlying Fig. 1b are provided as a Source Data file.