Abstract
Considerable efforts have been made to identify the genetic basis of human longevity, with only limited progress. One important drawback of current genetic studies is the limited knowledge of gene-environment interaction. Using 2 cohorts of long-lived individuals born in 1905 and 1915 in Denmark, we performed survival analysis to estimate risk of mortality for major candidate genes of aging and longevity and their cohort effects. Through statistical modeling that combines individual genetic and survival information with cohort-specific survival data, we estimated the relative risks of mortality from ages 95 to 103 years associated with genetic variants in apolipoprotein E (APOE), forkhead box class O3a, clusterin, and phosphatidylinositol binding clathrin assembly protein. Our analysis estimated a decreased risk of carrying the APOE 4 allele (change in risk = –0.403, 95% confidence interval (CI): −0.831, 0.021; P = 0.040) in men of the later cohort, although the allele itself was harmful to survival across sexes (relative risk = 1.161, 95% CI: 1.027, 1.345; P = 0.026). We also estimated a cohort effect of increased risk for the minor allele of rs3851179 in phosphatidylinositol binding clathrin assembly protein with borderline significance (change in risk = 0.165, 95% CI: −0.010, 0.331; P = 0.052) in women. Our estimated significant cohort effect on APOE
4 allele (change in risk = –0.403, 95% confidence interval (CI): −0.831, 0.021; P = 0.040) in men of the later cohort, although the allele itself was harmful to survival across sexes (relative risk = 1.161, 95% CI: 1.027, 1.345; P = 0.026). We also estimated a cohort effect of increased risk for the minor allele of rs3851179 in phosphatidylinositol binding clathrin assembly protein with borderline significance (change in risk = 0.165, 95% CI: −0.010, 0.331; P = 0.052) in women. Our estimated significant cohort effect on APOE 4 is indicative of the interplay between the gene and the changing environment that modulates survival at extreme ages.
4 is indicative of the interplay between the gene and the changing environment that modulates survival at extreme ages.
Keywords: advanced age, APOE, birth cohorts, CLU, FOXO3A, mortality, PICALM
Abbreviations
- AD
Alzheimer disease
- APOE
apolipoprotein E
- CLU
clusterin
- FOXO3A
forkhead box class O3a
- G × E
gene-environment interaction
- PICALM
phosphatidylinositol binding clathrin assembly protein
The concept of gene-environment interaction (G × E) refers to changes in the relative importance of the same genotype across different environmental conditions. G × E is likely a common phenomenon for traits or phenotypes under the control of genetic and environmental influences such as complex diseases (1) and longevity (2). The genetic contribution to human lifespan has been estimated to account for approximately one-fourth of the total lifespan variation in twin studies (3) or even lower in extended families (4) and after adjusting for assortative mating (5). The limited genetic contribution emphasizes the high importance of environmental factors that mediate variation in lifespan and longevity interactively with our genome whereby epigenetic remodeling might serve as an ideal mechanism for phenotypic plasticity.
Even though the estimated genetic contribution is relatively low, considerable efforts have been made in search of genetic variants associated with human longevity. In particular, high-throughput single-nucleotide polymorphism genotyping and modern sequencing methods (6) have enabled genome-wide association studies (GWAS) to identify genomic variants underlying human longevity (7–9). To date, the only consistent finding from GWAS of human longevity is the apolipoprotein E (APOE) gene, which was already discovered as a candidate gene decades ago (10) and has been estimated to account for no more than approximately 3.5% of the total variance in life expectancy (11). Another major longevity gene is forkhead box class O3a (FOXO3A), which has exhibited longevity association in diverse human populations (12–16). Other genes, such as clusterin (CLU) and phosphatidylinositol binding clathrin assembly protein (PICALM), have been associated with Alzheimer disease (AD) in GWAS (17, 18) and candidate gene studies (19–23). Given the fact that increasing age is the greatest known risk factor for AD, we assume that genes associated with AD might also correlate with survival at extreme age, possibly with cohort effects.
Based on genotype data from long-lived individuals from several Danish birth cohorts of the late 19th and early 20th century, we previously compared frequencies of APOE and FOXO3A variants across cohorts and reported a decrease in the allele frequencies of the APOE 4 allele and the FOXO3 rs7762395 minor allele in individuals from more recent birth cohorts, suggesting a genetic imprint of cohort differences in the selective pressure on surviving to high ages of over 95 years (24). The observed allele frequency differences across cohorts are indicative of G × E occurring for the 2 genes and their influences in living up to age 95, but they do not offer any cohort-related relative risk estimates for surviving at the more advanced ages after 95. By using genotype data for variants in APOE, FOXO3A, CLU, and PICALM on the same samples of 2 old Danish birth cohorts originally analyzed by Nygaard et al. (24) and applying a statistical model that combines cohort-specific life-table data with individual genotype data (25), we report our recently performed survival analysis to estimate the relative risk as well as cohort effects of genetic variants in major candidate genes of human aging and longevity on all-cause mortality at extreme ages from 95 to 103 years.
4 allele and the FOXO3 rs7762395 minor allele in individuals from more recent birth cohorts, suggesting a genetic imprint of cohort differences in the selective pressure on surviving to high ages of over 95 years (24). The observed allele frequency differences across cohorts are indicative of G × E occurring for the 2 genes and their influences in living up to age 95, but they do not offer any cohort-related relative risk estimates for surviving at the more advanced ages after 95. By using genotype data for variants in APOE, FOXO3A, CLU, and PICALM on the same samples of 2 old Danish birth cohorts originally analyzed by Nygaard et al. (24) and applying a statistical model that combines cohort-specific life-table data with individual genotype data (25), we report our recently performed survival analysis to estimate the relative risk as well as cohort effects of genetic variants in major candidate genes of human aging and longevity on all-cause mortality at extreme ages from 95 to 103 years.
METHODS
Study population
The study included long-lived individuals from the Danish 1905 and 1915 birth cohort studies. Details about the 2 studies are described elsewhere (24, 26). In brief, a total of 1,169 unrelated individuals born in 1905 (286 men and 883 women) and 1,105 unrelated individuals born in 1915 (302 men and 803 women) who consented to provide blood samples and survived past age 95 were included in the study. Individual survival status was retrieved from the Danish Central Population Register (27), with the vital status followed until death or January 1, 2018. Web Figure 1 (available at https://academic.oup.com/aje) presents Kaplan-Meier plots showing sex-specific survival curves for the 2 birth cohorts after age 95 years. The mean survival time for the 1905 birth cohort is 2 years for men and 3 years for women. For the 1915 birth cohort both men and women have a mean survival time of 3 years although men display a clear pattern of lower survival probability at later ages (sex difference in mean survival time of <0.5 years). As shown in Web Figure 1, the 1915 birth cohort is truncated at age 104 (9 years after age 95) due to the latest mortality update in 2018. With that consideration, our model fitting is restricted to an age range of 95–103 years for both birth cohorts.
For each blood sample, extracted DNA was analyzed for haplotype groups ( 2,
2,  3,
3,  4) in APOE, rs479744 and rs7762395 in FOXO3A, rs11136000 in CLU, and rs3851179 in PCALM. Details of the genotyping experiment and quality control have been described in Nygaard et al. (24). Permission to collect blood samples and use of register-based information was granted by the Danish Scientific-Ethical Committees.
4) in APOE, rs479744 and rs7762395 in FOXO3A, rs11136000 in CLU, and rs3851179 in PCALM. Details of the genotyping experiment and quality control have been described in Nygaard et al. (24). Permission to collect blood samples and use of register-based information was granted by the Danish Scientific-Ethical Committees.
Statistical analysis
Based on the observed individual genotypes and the recently updated mortality information available for the 2 birth cohorts, we conducted a semiparametric survival analysis by introducing cohort- and sex-specific population survivals from population statistics and specifying and estimating genotype-specific relative risks in the 1905 birth cohort and change in relative risk from 1905–1915 with
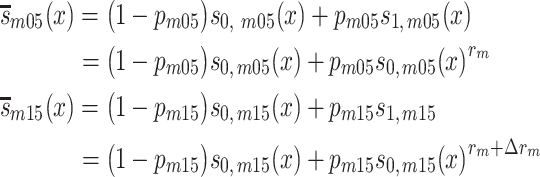 |
(1) |
for men, and
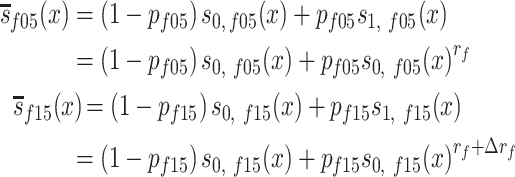 |
(2) |
for women. In equations (1) and (2), 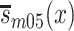 and
and 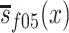 are population survival functions for men and women from the 1905 birth cohort conditioned on surviving to age 95 years. Likewise,
are population survival functions for men and women from the 1905 birth cohort conditioned on surviving to age 95 years. Likewise, 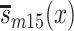 and
and 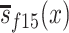 are sex-specific conditional survival functions for the 1915 birth cohort.
are sex-specific conditional survival functions for the 1915 birth cohort.  and
and 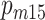 are frequencies of male carriers of a genetic variant in the 1905 and 1915 birth cohorts at age 95, whereas
are frequencies of male carriers of a genetic variant in the 1905 and 1915 birth cohorts at age 95, whereas 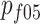 and
and 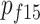 are cohort-specific carrier frequencies in women at age 95.
are cohort-specific carrier frequencies in women at age 95.  ,
, 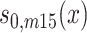 ,
,  , and
, and  are sex- and cohort-specific baseline survival function for noncarriers, and
are sex- and cohort-specific baseline survival function for noncarriers, and 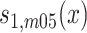 ,
,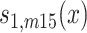 ,
, 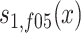 , and
, and 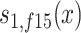 are sex- and cohort-specific survival functions for carriers; rm and rf are sex-specific relative risks of survival in carriers, and ∆rm and ∆rf are the changes in relative risk from the 1905 to the 1915 birth cohort. With equations (1) and (2), we can calculate, for each sex and cohort, the proportion of carriers at age x (x ≥ 95) as
are sex- and cohort-specific survival functions for carriers; rm and rf are sex-specific relative risks of survival in carriers, and ∆rm and ∆rf are the changes in relative risk from the 1905 to the 1915 birth cohort. With equations (1) and (2), we can calculate, for each sex and cohort, the proportion of carriers at age x (x ≥ 95) as
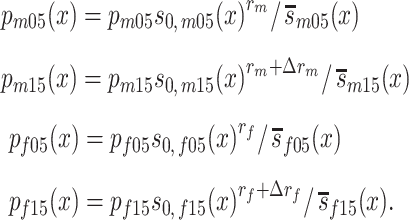 |
(3) |
Because the genotype frequencies over ages in equation (3) are from the same cohort, parameter estimation should take into account related observations. Here, we can estimate rm and ∆rm for men and rf and ∆rf for women using an optimization approach that minimizes the absolute difference between the estimated and observed age-specific genotype frequency in male and female samples. Significance of the parameter estimates was obtained by a randomization test using 1,000 permutations and 95% confidence intervals assessed by bootstrapping using 1,000 replicates.
In equations (1) and (2), the genotype-specific survival functions can be modified to take into consideration unobserved frailty. Taking a closer look at the men from the 1905 birth cohort for example, we have.
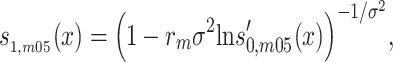 |
(4) |
where 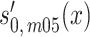 is the baseline survival function in a homogeneous population and
is the baseline survival function in a homogeneous population and  2 the variance of unobserved frailty (set to 0.1) (28). The same replacement can be done for men from the 1915 birth cohort and for women from both cohorts for fitting frailty models.
2 the variance of unobserved frailty (set to 0.1) (28). The same replacement can be done for men from the 1915 birth cohort and for women from both cohorts for fitting frailty models.
Assuming no sex-specific genetic influence, we can replace the sex-specific cohort survival with sex-combined cohort survival for parameter estimation. The sex-combined and sex-specific survival functions for the 1905 and 1915 birth cohorts were taken from the mortality database at http://www.mortality.org/, hosted by the Max-Planck Institute for Demographic Research (Rostock, Germany).
Statistical analysis was performed using the Gauss platform (https://www.aptech.com/) for semiparametric survival analysis and using R (R Foundation for Statistical Computing, Vienna, Austria) for Cox regression analysis and graphics.
RESULTS
In Table 1, we show the observed frequencies of carriers of APOE haplotype groups and minor single-nucleotide polymorphism alleles in FOXO3A, CLU, and PICALM genes. The observed carrier frequencies support the trends of genotypes for APOE and FOXO3A as reported by Nygaard et al. (24), although statistical tests (χ2 test) revealed no significant cohort difference in the carrier frequencies.
Table 1.
Observations and Frequency of Carriers of Selected Gene Variants, According to Sex and Birth Cohorts, Denmark, Initiated in 1998 (1905 Cohort) and 2010 (1915 Cohort)
| Men | Women | All | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1905 | 1915 | 1905 | 1915 | 1905 | 1915 | ||||||||||
| Genetic Variant | Obs a | Freq | Obs | Freq | P Value | Obs | Freq | Obs | Freq | P Value | Obs | Freq | Obs | Freq | P Value |
| APOE | |||||||||||||||
 2 2 |
285 | 0.182 | 301 | 0.246 | 0.077 | 878 | 0.198 | 803 | 0.215 | 0.416 | 1,163 | 0.194 | 1,104 | 0.224 | 0.095 |
 4 4 |
285 | 0.218 | 301 | 0.243 | 0.536 | 878 | 0.216 | 803 | 0.183 | 0.100 | 1,163 | 0.217 | 1,104 | 0.199 | 0.333 |
| FOXO3A | |||||||||||||||
| rs479744 | 268 | 0.381 | 301 | 0.336 | 0.302 | 833 | 0.375 | 803 | 0.374 | 1.000 | 1,101 | 0.376 | 1,104 | 0.363 | 0.563 |
| rs7762395 | 278 | 0.291 | 301 | 0.243 | 0.217 | 841 | 0.271 | 803 | 0.291 | 0.390 | 1,119 | 0.276 | 1,104 | 0.278 | 0.956 |
| CLU | |||||||||||||||
| rs11136000 | 285 | 0.593 | 301 | 0.628 | 0.434 | 880 | 0.606 | 803 | 0.634 | 0.255 | 1,165 | 0.603 | 1,104 | 0.632 | 0.159 |
| PICALM | |||||||||||||||
| rs3851179 | 284 | 0.630 | 301 | 0.611 | 0.698 | 874 | 0.628 | 803 | 0.618 | 0.696 | 1,158 | 0.629 | 1,104 | 0.616 | 0.561 |
Abbreviations: APOE, apolipoprotein E; CLU, clusterin; FOXO3A, forkhead box class O3a; Freq, frequency; Obs, observations; PICALM, phosphatidylinositol binding clathrin assembly protein.
a Obs: total number of carriers and noncarriers of an APOE allele or of the minor allele of a single nucleotide polymorphism.
We first fitted semiparametric survival models to the APOE haplotypes ( 2,
2,  4). As shown in Table 2, the models estimated significant relative risks in the sex-combined samples for the
4). As shown in Table 2, the models estimated significant relative risks in the sex-combined samples for the  4 allele (no-frailty model: r = 1.161, P = 0.026; frailty model: r = 1.183, P = 0.020). In the sex-specific models, no significant risk was estimated although both sexes had similar influence of increased hazard of death. Importantly, the sex-specific model estimated a significant cohort effect for the
4 allele (no-frailty model: r = 1.161, P = 0.026; frailty model: r = 1.183, P = 0.020). In the sex-specific models, no significant risk was estimated although both sexes had similar influence of increased hazard of death. Importantly, the sex-specific model estimated a significant cohort effect for the  4 allele in men (no-frailty model: ∆r = −0.403, P = 0.040; frailty model: ∆r = −0.466, P = 0.044). The relative risk for carrying
4 allele in men (no-frailty model: ∆r = −0.403, P = 0.040; frailty model: ∆r = −0.466, P = 0.044). The relative risk for carrying  2 is not different from 1 and their cohort effects are not different from 0. Figure 1 displays the observed and estimated frequencies of
2 is not different from 1 and their cohort effects are not different from 0. Figure 1 displays the observed and estimated frequencies of  4 allele carriers by age in male (Figure 1A and 1D), female (Figure 1B and 1E), and sex-combined (Figure 1C and 1F) samples of the 2 birth cohorts. Notably, the male carriers exhibit opposite age patterns indicating reduced or reversed influences of the
4 allele carriers by age in male (Figure 1A and 1D), female (Figure 1B and 1E), and sex-combined (Figure 1C and 1F) samples of the 2 birth cohorts. Notably, the male carriers exhibit opposite age patterns indicating reduced or reversed influences of the  4 allele on survival at extreme ages.
4 allele on survival at extreme ages.
Table 2.
Estimated Relative Risk r and Its Cohort Difference ∆r for Apolipoprotein E Gene Variants, Using 2 Birth Cohorts and Their Updated Cohort-Specific Survival Data, in Denmark, as of 2018
| Men | Women | All | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Risk Parameter | Median | 95% CI | P Value | Median | 95% CI | P Value | Median | 95% CI | P Value |
APOE
 2 2 |
|||||||||
| No frailty | |||||||||
| r | 1.098 | 0.799, 1.386 | 0.514 | 0.991 | 0.858, 1.162 | 0.926 | 1.004 | 0.885, 1.157 | 0.992 |
| ∆r | −0.103 | −0.483, 0.375 | 0.650 | 0.000 | −0.215, 0.202 | 0.988 | −0.003 | −0.195, 0.163 | 0.958 |
| Frailty | |||||||||
| r | 1.109 | 0.795, 1.481 | 0.612 | 0.991 | 0.855, 1.199 | 0.942 | 1.004 | 0.874, 1.174 | 1.000 |
| ∆r | −0.113 | −0.512, 0.422 | 0.594 | −0.001 | −0.239, 0.231 | 0.964 | −0.004 | −0.208, 0.183 | 0.938 |
APOE
 4 4 |
|||||||||
| No frailty | |||||||||
| r | 1.276 | 0.972, 1.651 | 0.102 | 1.135 | 0.986, 1.354 | 0.088 | 1.161 | 1.027, 1.345 | 0.026 |
| ∆r | −0.403 | −0.831, −0.021 | 0.040 | −0.116 | −0.394, 0.105 | 0.306 | −0.156 | −0.364, 0.030 | 0.086 |
| Frailty | |||||||||
| r | 1.325 | 0.993, 1.796 | 0.102 | 1.154 | 0.980, 1.414 | 0.068 | 1.183 | 1.014, 1.385 | 0.020 |
| ∆r | −0.466 | −1.003, −0.002 | 0.044 | −0.131 | −0.441, 0.112 | 0.280 | −0.177 | −0.412, 0.035 | 0.076 |
Abbreviations: APOE, apolipoprotein E; CI, confidence interval.
Figure 1.

The observed (dash-circle line) and estimated (dash-dot line, no frailty) frequency of apolipoprotein E  4 allele carriers, Denmark, 1905 and 1915 birth cohorts. In the 1905 birth cohort: men (A), women (B), and both combined (C); in the 1915 birth cohort: men (D), women (E), and both combined (F).
4 allele carriers, Denmark, 1905 and 1915 birth cohorts. In the 1905 birth cohort: men (A), women (B), and both combined (C); in the 1915 birth cohort: men (D), women (E), and both combined (F).
Table 3 presents the parameter estimates for the rest of the investigated candidate genes. No variant in the genes showed significant estimates—neither for relative risk, r, nor for cohort effect, ∆r. However, female carriers of the minor allele of rs3851179 in PICALM tended to have increased risk of death in the later cohort with borderline significance (no-frailty model: ∆r = 0.165, P = 0.052; frailty model: ∆r = 0.185, P = 0.058). The observed and estimated frequencies of rs3851179 minor allele carriers according to age are shown in Figure 2 for male (Figure 2A and 2D), female (Figure 2B and 2E), and sex-combined (Figure 2C and 2F) samples of the 2 birth cohorts. In Figure 2, the female age pattern of the observed and fitted frequencies of minor allele carriers increased in the 1905 cohort (Figure 2B) but decreased in the 1915 cohort (Figure 2E).
Table 3.
Estimated Relative Risk r and Its Cohort Difference ∆r for Selected Candidate Genes Using 2 Birth Cohorts and Their Updated Cohort-Specific Survival Data, in Denmark, as of 2018
| Men | Women | All | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Risk Parameter | Median | 95% CI | P Value | Median | 95% CI | P Value | Median | 95% CI | P Value |
| FOXO3A rs479744 | |||||||||
| No frailty | |||||||||
| r | 0.887 | 0.692, 1.136 | 0.456 | 1.013 | 0.892, 1.158 | 0.874 | 0.987 | 0.882, 1.115 | 0.772 |
| ∆r | 0.160 | −0.162, 0.532 | 0.382 | 0.067 | −0.109, 0.240 | 0.414 | 0.076 | −0.082, 0.230 | 0.318 |
| Frailty | |||||||||
| r | 0.872 | 0.645, 1.164 | 0.486 | 1.014 | 0.878, 1.174 | 0.812 | 0.984 | 0.874, 1.126 | 0.780 |
| ∆r | 0.179 | −0.205, 0.589 | 0.380 | 0.078 | −0.135, 0.269 | 0.428 | 0.087 | −0.090, 0.271 | 0.318 |
| FOXO3A rs7762395 | |||||||||
| No frailty | |||||||||
| r | 0.904 | 0.708, 1.201 | 0.536 | 1.113 | 0.970, 1.284 | 0.108 | 1.064 | 0.944, 1.205 | 0.292 |
| ∆r | 0.099 | −0.270, 0.425 | 0.604 | −0.057 | −0.261, 0.139 | 0.544 | −0.034 | −0.198, 0.125 | 0.696 |
| Frailty | |||||||||
| r | 0.892 | 0.662, 1.237 | 0.492 | 1.128 | 0.976, 1.339 | 0.096 | 1.072 | 0.938, 1.231 | 0.292 |
| ∆r | 0.108 | −0.298, 0.497 | 0.652 | −0.063 | −0.303, 0.151 | 0.540 | −0.038 | −0.223, 0.163 | 0.650 |
| CLU rs11136000 | |||||||||
| No frailty | |||||||||
| r | 0.912 | 0.685, 1.168 | 0.134 | 1.065 | 0.946, 1.213 | 0.272 | 1.029 | 0.917, 1.146 | 0.558 |
| ∆r | −0.104 | −0.422, 0.156 | 0.119 | −0.043 | −0.210, 0.126 | 0.628 | −0.044 | −0.197, 0.104 | 0.554 |
| Frailty | |||||||||
| r | 0.902 | 0.663, 1.160 | 0.434 | 1.073 | 0.934, 1.218 | 0.258 | 1.033 | 0.918, 1.163 | 0.536 |
| ∆r | −0.117 | −0.490, 0.237 | 0.534 | −0.047 | −0.247, 0.135 | 0.650 | −0.049 | −0.209, 0.115 | 0.606 |
| PICALM rs3851179 | |||||||||
| No frailty | |||||||||
| r | 0.954 | 0.736, 1.196 | 0.654 | 0.951 | 0.829, 1.071 | 0.372 | 0.954 | 0.849, 1.063 | 0.354 |
| ∆r | −0.028 | −0.351, 0.310 | 0.864 | 0.165 | −0.010, 0.331 | 0.052 | 0.127 | −0.014, 0.273 | 0.082 |
| Frailty | |||||||||
| r | 0.944 | 0.692, 1.220 | 0.696 | 0.947 | 0.796, 1.091 | 0.396 | 0.949 | 0.836, 1.076 | 0.344 |
| ∆r | −0.027 | −0.392, 0.345 | 0.836 | 0.185 | −0.012, 0.385 | 0.058 | 0.143 | −0.029, 0.322 | 0.098 |
Abbreviations: CI, confidence interval; CLU, clusterin; FOXO3A, forkhead box class O3a; PICALM, phosphatidylinositol binding clathrin assembly protein.
Figure 2.

The observed (dash-circle line) and estimated (dash-dot line, no frailty) frequency of carriers of forkhead box class O3a rs479744 minor allele, Denmark, 1905 and 1915 birth cohorts. In the 1905 birth cohort: men (A), women (B), and both combined (C); in the 1915 birth cohort: men (D), women (E), and both combined (F).
For comparison purposes, we fitted Cox proportional hazard models to the data on APOE 4 and on rs3851179 in the PICALM gene, and the results are shown in Web Tables 1 and 2. The Cox model estimated marginal risks of genotype, cohort, and their G × E equivalent to the cohort difference in the risk of genotype in our semiparametric model. The Cox model estimated a significant risk for carrying the APOE
4 and on rs3851179 in the PICALM gene, and the results are shown in Web Tables 1 and 2. The Cox model estimated marginal risks of genotype, cohort, and their G × E equivalent to the cohort difference in the risk of genotype in our semiparametric model. The Cox model estimated a significant risk for carrying the APOE 4 allele in the sex-combined sample surprisingly with nearly similar effect size (hazard ratio = 1.178) and P value (0.022) as in our semiparametric model (no-frailty model: r = 1.161, P value = 0.026; frailty model: r = 1.183, P value = 0.020). Likewise, both the Cox model and the semiparametric model produced comparable estimates on the risk of death for carrying the APOE
4 allele in the sex-combined sample surprisingly with nearly similar effect size (hazard ratio = 1.178) and P value (0.022) as in our semiparametric model (no-frailty model: r = 1.161, P value = 0.026; frailty model: r = 1.183, P value = 0.020). Likewise, both the Cox model and the semiparametric model produced comparable estimates on the risk of death for carrying the APOE 4 allele in male and female samples (Table 2, Web Table 1). However, no significant G × E was estimated by the Cox model fitted to the male samples, although the model also estimated a reduced influence of the
4 allele in male and female samples (Table 2, Web Table 1). However, no significant G × E was estimated by the Cox model fitted to the male samples, although the model also estimated a reduced influence of the  4 allele in the later cohort, and its P value was the lowest (0.135) for G × E in the sex-specific and sex-combined analyses. No significant influence was estimated except for the marginal risk of cohort irrelevant to the genotype of PICALM (Web Table 2).
4 allele in the later cohort, and its P value was the lowest (0.135) for G × E in the sex-specific and sex-combined analyses. No significant influence was estimated except for the marginal risk of cohort irrelevant to the genotype of PICALM (Web Table 2).
Finally, we performed a replication analysis of the significant finding on the APOE 4 allele using the other small cohorts reported in Nygaard et al. (24), that is, the 1895 (125 individuals), 1910 (176 individuals), and 1911 (130 individuals) birth cohorts. Altogether we had 431 individuals (103 men and 328 women) with ages ranging from 100 to 109 years. Because of the small sample sizes, especially after dividing by sex, we were unable to fit the semiparametric model but fitted the simple Cox regression models instead. In the Cox model, we included APOE
4 allele using the other small cohorts reported in Nygaard et al. (24), that is, the 1895 (125 individuals), 1910 (176 individuals), and 1911 (130 individuals) birth cohorts. Altogether we had 431 individuals (103 men and 328 women) with ages ranging from 100 to 109 years. Because of the small sample sizes, especially after dividing by sex, we were unable to fit the semiparametric model but fitted the simple Cox regression models instead. In the Cox model, we included APOE 4 genotype (carrier coded 1; noncarrier coded 0), cohort year, and their interaction. Here the interaction term estimates the genotype influence conditional on the cohort difference. All models estimated increased hazard of death for carrying the APOE
4 genotype (carrier coded 1; noncarrier coded 0), cohort year, and their interaction. Here the interaction term estimates the genotype influence conditional on the cohort difference. All models estimated increased hazard of death for carrying the APOE 4 allele, but none was significant. Interestingly, the sex-combined analysis estimated a reduced hazard of death for later cohorts (hazard ratio = 0.986, 95% confidence interval: 0.971, 1.002; P = 0.078). Importantly, the interaction term representing cohort effect of APOE
4 allele, but none was significant. Interestingly, the sex-combined analysis estimated a reduced hazard of death for later cohorts (hazard ratio = 0.986, 95% confidence interval: 0.971, 1.002; P = 0.078). Importantly, the interaction term representing cohort effect of APOE 4 on survival had a hazard ratio of <1 in all models in support of our finding in 1905 and 1915 birth cohorts, although no model had a P value of <0.05. In the sex-combined analysis, the hazard ratio for interaction was 0.977 (95% confidence interval: 0.942, 1.013; P = 0.205). For sex-specific analyses, we obtained a hazard ratio of 0.987 (95% confidence interval: 0.946, 1.030; P = 0.557) for women and a hazard ratio of 0.947 (95% confidence interval: 0.884, 1.014; P = 0.119) for men, with the men’s result having the lowest P value. The pattern is in good agreement with our estimates in Table 2.
4 on survival had a hazard ratio of <1 in all models in support of our finding in 1905 and 1915 birth cohorts, although no model had a P value of <0.05. In the sex-combined analysis, the hazard ratio for interaction was 0.977 (95% confidence interval: 0.942, 1.013; P = 0.205). For sex-specific analyses, we obtained a hazard ratio of 0.987 (95% confidence interval: 0.946, 1.030; P = 0.557) for women and a hazard ratio of 0.947 (95% confidence interval: 0.884, 1.014; P = 0.119) for men, with the men’s result having the lowest P value. The pattern is in good agreement with our estimates in Table 2.
DISCUSSION
Based on 2 unique birth cohorts of long-lived individuals, we were able to estimate the relative risk as well as cohort effect of variants in major candidate genes of human aging and longevity by performing survival analysis that combines individual genotype and survival information with cohort-specific life-table data. Whereas our analysis supports previous conclusions concerning the negative association of the APOE 4 allele with human longevity (7, 29, 30), our strategic modeling on the 2 cohorts was, we believe for the first time, able to estimate the change over time in the influences on all-cause mortality by major aging and longevity candidate genes.
4 allele with human longevity (7, 29, 30), our strategic modeling on the 2 cohorts was, we believe for the first time, able to estimate the change over time in the influences on all-cause mortality by major aging and longevity candidate genes.
Among the multiple candidate genes, only the APOE  4 allele showed significant estimates on its overall risk in sex-combined samples and cohort effect in the male-only samples, with the same trend replicated by independent data in other birth cohorts. Although the estimated cohort effect of the
4 allele showed significant estimates on its overall risk in sex-combined samples and cohort effect in the male-only samples, with the same trend replicated by independent data in other birth cohorts. Although the estimated cohort effect of the  4 allele is significant only for men, the same direction of influence was also estimated in female samples. In Figure 1, the age patterns in women are not strikingly different from those in men, but we do observe a slower rate of change of carrier frequency over ages in the 1915 as compared with the 1905 birth cohort. This result seems contradictory to our previous report of reduced proportion of
4 allele is significant only for men, the same direction of influence was also estimated in female samples. In Figure 1, the age patterns in women are not strikingly different from those in men, but we do observe a slower rate of change of carrier frequency over ages in the 1915 as compared with the 1905 birth cohort. This result seems contradictory to our previous report of reduced proportion of  4 allele carriers in the 1915 cohort, possibly ascribable to an increased risk of the allele on mortality (24). However, it needs to be stated that the published result was on the overall impact of the
4 allele carriers in the 1915 cohort, possibly ascribable to an increased risk of the allele on mortality (24). However, it needs to be stated that the published result was on the overall impact of the  4 allele on surviving from birth to past age 95 years, while our estimate is for the influence in surviving to 95–103 years, that is, survival at extreme ages. Putting it all together, although our estimate shows that the
4 allele on surviving from birth to past age 95 years, while our estimate is for the influence in surviving to 95–103 years, that is, survival at extreme ages. Putting it all together, although our estimate shows that the  4 allele increases the risk of death at extreme ages, its risk on mortality at advanced ages is slightly reduced in the more recent cohort, although significantly only in men. Note that an increased risk of death at more advanced ages in carriers of the APOE
4 allele increases the risk of death at extreme ages, its risk on mortality at advanced ages is slightly reduced in the more recent cohort, although significantly only in men. Note that an increased risk of death at more advanced ages in carriers of the APOE 4 allele was found for the 1905 birth cohort (28, 30), which seems inconsistent with the present finding. In that respect it should be emphasized that the present estimate of reduced influence is on cohort differences in the risk of the
4 allele was found for the 1905 birth cohort (28, 30), which seems inconsistent with the present finding. In that respect it should be emphasized that the present estimate of reduced influence is on cohort differences in the risk of the  4 allele, while the previous estimate of increased risk with advancing ages was on the same cohort.
4 allele, while the previous estimate of increased risk with advancing ages was on the same cohort.
Previous studies have associated rs3851179 in the PICALM gene with AD (21–23), with reduced risk from its minor allele. Interestingly, our estimated marginal influences on survival for minor allele carriers are all lower than 1, suggesting its negative (although statistically insignificant) correlation with mortality at advanced ages. The borderline significance of its cohort effect in women could, on the other hand, suggest that its impact at extreme ages might have been reversed by the changing environment in a sex-specific manner. Comparing the sex-specific age patterns of the 1915 birth cohort in Figure 2, it is clear that the same polymorphism was expressed very differently in men (protective) and women (harmful) in terms of survival. This interesting finding, although only borderline significant, merits further investigation, replication, and validation.
The 10-year difference in the cohorts’ birth years (from 1905–1915) should not be the sole reason for the estimated cohort effect. In fact, the 2 cohorts experienced almost the same environmental conditions for around a century but with a 10-year shift in their ages of exposure. That is, the latter cohort benefited from almost the same environmental improvements (socioeconomic and medical) as the earlier cohort, but 10 years earlier, for nearly a century. We know that the past 100 years encompass the third industrial revolution, with the rise of electronics, and currently the fourth industrial revolution, with the emergence of the internet, both accompanied by remarkable advancements in medicine and improvements in treatment and health care. We could postulate that the estimated cohort effect reflects a cumulative influence of earlier exposure to the improving environments by the later cohort.
It is interesting that both the Cox model and our semiparametric model gave approximately similar risk estimates for carrying the APOE 4 allele in the sex-combined sample (Cox model, hazard ratio = 1.178, P value: 0.022; semiparametric model, relative risk = 1.161, P value of 0.026, in the no-frailty model and r = 1.183, P value of 0.020, in the frailty model) and in sex-specific samples, although not statistically significant in both models (P value < 0.1). However, the Cox model failed to detect the G × E corresponding to the cohort effect in the semiparametric model, although the Cox model estimated a reduced risk of the
4 allele in the sex-combined sample (Cox model, hazard ratio = 1.178, P value: 0.022; semiparametric model, relative risk = 1.161, P value of 0.026, in the no-frailty model and r = 1.183, P value of 0.020, in the frailty model) and in sex-specific samples, although not statistically significant in both models (P value < 0.1). However, the Cox model failed to detect the G × E corresponding to the cohort effect in the semiparametric model, although the Cox model estimated a reduced risk of the  4 allele in the later cohort in both sex-specific and sex-combined models (Web Table 1). Overall, the Cox model could be used to capture the marginal influence for genotype but failed in estimating the cohort difference in the influence of genotype, presumably due to the fact that the Cox model assumed a proportional hazard between the 2 cohorts which might deviate from the observed patterns used by semiparametric model.
4 allele in the later cohort in both sex-specific and sex-combined models (Web Table 1). Overall, the Cox model could be used to capture the marginal influence for genotype but failed in estimating the cohort difference in the influence of genotype, presumably due to the fact that the Cox model assumed a proportional hazard between the 2 cohorts which might deviate from the observed patterns used by semiparametric model.
In our analysis, both no-frailty and frailty models were fitted. As can be seen in Tables 2 and 3, the estimated risks are predominantly higher when using the frailty model. Although conventional survival analysis assumes a homogeneous population, just as in our no-frailty model, the estimates achieved when using the frailty model should be more realistic because a completely homogeneous population is hard to imagine. Note, however, that both models assume proportional hazards such that the age-dependent influence on mortality is ignored in the studied age range. Considering that our estimates were made over a relatively short age range of 95–103 years, the proportional hazards assumption seems reasonably acceptable.
The rapid decrease in sample size by age, as shown by the numbers in Figures 1 and 2, limits the statistical power of our analysis. Although the semiparametric model is advantaged by making use of the cohort-specific population survival data, the most significant estimate on cohort effect was only at a P value of 0.04. Moreover, the numbers in the figures also show the large difference in male and female sample sizes. Because of that, the sex-combined results are dominated mainly by the female samples. As a result, the overall conclusions of insignificant estimate for ∆r can be ascribed to the high proportion of women in the sex-combined analysis.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Epidemiology, Biostatistics and Biodemography, Department of Public Health, University of Southern Denmark, Odense C, Denmark (Qihua Tan, Rune Jacobsen, Marianne Nygaard, Mette Soerensen, Jonas Mengel-From, Kaare Christensen); Unit of Human Genetics, Department of Clinical Research, University of Southern Denmark, Odense C, Denmark (Qihua Tan, Kaare Christensen); and Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark (Lene Christiansen).
This work was funded jointly by the National Institutes of Health/National Institute on Aging (grant P01 AG08761) and Velux Foundation (research grant 00012540).
We thank Lisbeth Aagaard Larsen for providing the newest update of mortality data on the 1905 and 1915 birth cohorts.
Conflict of interest: none declared.
REFERENCES
- 1. Dempfle A, Scherag A, Hein R, et al. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16(10):1164–1172. [DOI] [PubMed] [Google Scholar]
- 2. Dato S, Rose G, Crocco P, et al. The genetics of human longevity: an intricacy of genes, environment, culture and microbiome. Mech Ageing Dev. 2017;165(B):147–155. [DOI] [PubMed] [Google Scholar]
- 3. Herskind AM, McGue M, Iachine IA, et al. Untangling genetic influences on smoking, body mass index and longevity: a multivariate study of 2464 Danish twins followed for 28 years. Hum Genet. 1996;98(4):467–475. [DOI] [PubMed] [Google Scholar]
- 4. Kaplanis J, Gordon A, Shor T, et al. Quantitative analysis of population-scale family trees with millions of relatives. Science. 2018;360(6385):171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruby JG, Wright KM, Rand KA, et al. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics. 2018;210(3):1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deelen J, Beekman M, Capri M, et al. Identifying the genomic determinants of aging and longevity in human population studies: progress and challenges. Bioessays. 2013;35(4):386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deelen J, Beekman M, Uh HW, et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet. 2014;23(16):4420–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng Y, Nie C, Min J, et al. Novel loci and pathways significantly associated with longevity. Sci Rep. 2016;6:21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schächter F, Faure-Delanef L, Guénot F, et al. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6(1):29–32. [DOI] [PubMed] [Google Scholar]
- 11. Ewbank DC. The APOE gene and differences in life expectancy in Europe. J Gerontol A Biol Sci Med Sci. 2004;59(1):16–20. [DOI] [PubMed] [Google Scholar]
- 12. Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105(37):13987–13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anselmi CV, Malovini A, Roncarati R, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12(2):95–104. [DOI] [PubMed] [Google Scholar]
- 14. Soerensen M, Dato S, Christensen K, et al. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9(6):1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng Y, Cheng L, Chen H, et al. Effects of FOXO genotypes on longevity: a biodemographic analysis. J Gerontol A Biol Sci Med Sci. 2010;65(12):1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris BJ, Willcox DC, Donlon TA, et al. FOXO3: a major gene for human longevity—a mini-review. Gerontology. 2015;61(6):515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. [DOI] [PubMed] [Google Scholar]
- 19. Mengel-From J, Christensen K, McGue M, et al. Genetic variations in the CLU and PICALM genes are associated with cognitive function in the oldest old. Neurobiol Aging. 2010;32(3):554.e7–554.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corneveaux JJ, Myers AJ, Allen AN, et al. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19(16):3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kok EH, Luoto T, Haikonen S, et al. CLU, CR1 and PICALM genes associate with Alzheimer's-related senile plaques. Alzheimers Res Ther. 2011;3(2):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu G, Zhang S, Cai Z, et al. PICALM gene rs3851179 polymorphism contributes to Alzheimer's disease in an Asian population. Neuromolecular Med. 2013;15(2):384–388. [DOI] [PubMed] [Google Scholar]
- 23. Santos-Rebouças CB, Gonçalves AP, Dos Santos JM, et al. rs3851179 polymorphism at 5′ to the PICALM gene is associated with Alzheimer and Parkinson diseases in Brazilian population. Neuromolecular Med. 2017;19(2–3):293–299. [DOI] [PubMed] [Google Scholar]
- 24. Nygaard M, Lindahl-Jacobsen R, Soerensen M, et al. Birth cohort differences in the prevalence of longevity-associated variants in APOE and FOXO3A in Danish long-lived individuals. Exp Gerontol. 2014;57:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yashin AI, De Benedictis G, Vaupel JW, et al. Genes, demography, and life span: the contribution of demographic data in genetic studies on aging and longevity. Am J Hum Genet. 1999;65(4):1178–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasmussen SH, Andersen-Ranberg K, Thinggaard M, et al. Cohort profile: the 1895, 1905, 1910 and 1915 Danish Birth Cohort studies—secular trends in the health and functioning of the very old. Int J Epidemiol. 2017;46(6):1746–1746j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pedersen CB, Gøtzsche H, Møller JO, et al. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53(4):441–449. [PubMed] [Google Scholar]
- 28. Tan Q, Jacobsen R, Sørensen M, et al. Analyzing age-specific genetic effects on human extreme age survival in cohort-based longitudinal studies. Eur J Hum Genet. 2013;21(4):451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan Q, Christiansen L, Christensen K, et al. Apolipoprotein E genotype frequency patterns in aged Danes as revealed by logistic regression models. Eur J Epidemiol. 2004;19(7):651–656. [DOI] [PubMed] [Google Scholar]
- 30. Jacobsen R, Martinussen T, Christiansen L, et al. Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell. 2010;9(6):1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


