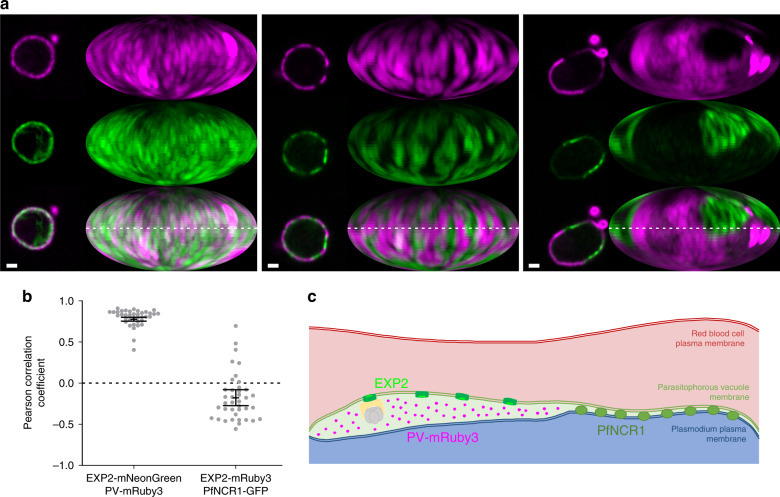
Fig. 4. Anti-colocalization of EXP2 and PfNCR1.
a Center confocal slice and Mollweide projections of PfNCR1-GFP—EXP2-mRuby3 parasites. EXP2-mRuby3 (top, magenta), PfNCR1-GFP (middle, green), merge (bottom). Samples chosen represent examples of relatively low (0.48), average (−0.09) and high (−0.55) anti-correlation according to the Pearson correlation coefficients of the maps. The dotted line labels the equator, corresponds to the confocal slice. Scale bar: 1 µm. b Pearson correlation coefficients of EXP2-mNeonGreen—PV-mRuby3 parasites (N = 38 cells, see Fig. 3) in comparison to PfNCR1-GFP—EXP2-mRuby3 (N = 39 cells). Regions that extend from the parasite to form the tubulovesicular network were excluded for the correlation analysis. Bars: mean ± 95% CI. Both distributions are significantly different: P = 1.2 × 10−29 (t-test, two tailed). c Localization of EXP2 correlates with the existence of a relatively large PV lumen. The PV lumen can store proteins for export and accessory proteins to facilitate protein export. The lipid transporter PfNCR1 localizes to regions of close PPM-PVM apposition. Functionally, PfNCR1 may be able to directly contact the PVM to exchange lipids and sites of membrane apposition may be sites for the general exchange of lipophilic material, a functional hallmark of membrane contact sites. Source data are provided as a Source data file.