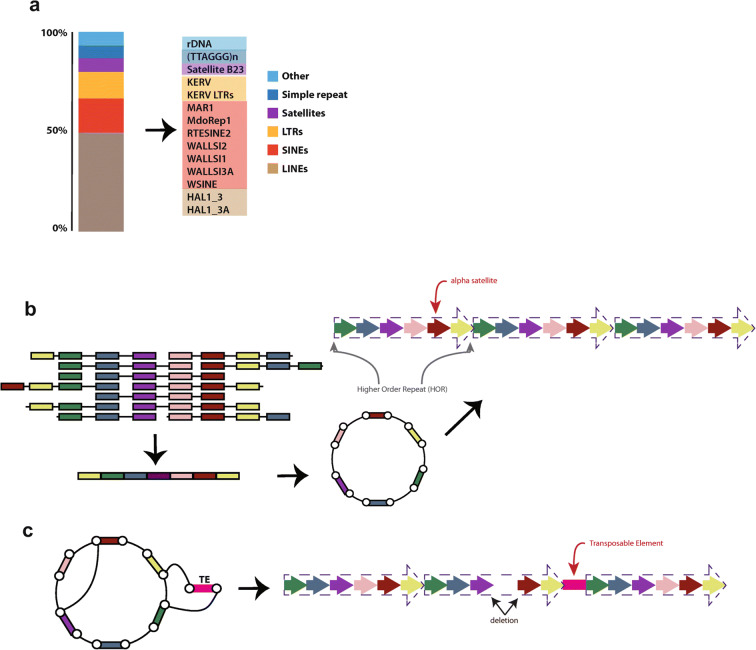
Fig. 2.
Examples of methods used to study repeats in the absence of a genome assembly. a Repeat Masker applied to raw sequencing data provides details on overall frequency of repeats by class (left) and specific type (right). c The linear order of highly repeated sequences, such as human alpha satellites found in centromeres, can be inferred from whole genome shotgun data (paired end sequencing). The resulting graphical model illustrates the frequency and order of satellite sequences (colored blocks). From the circular model, a linear arrangement of centromere satellites (colored arrows) can be inferred, including higher order repeat arrays (dotted arrows). c Variations within centromere arrays, such as deletions and insertions, can be captured with the graphical model approach