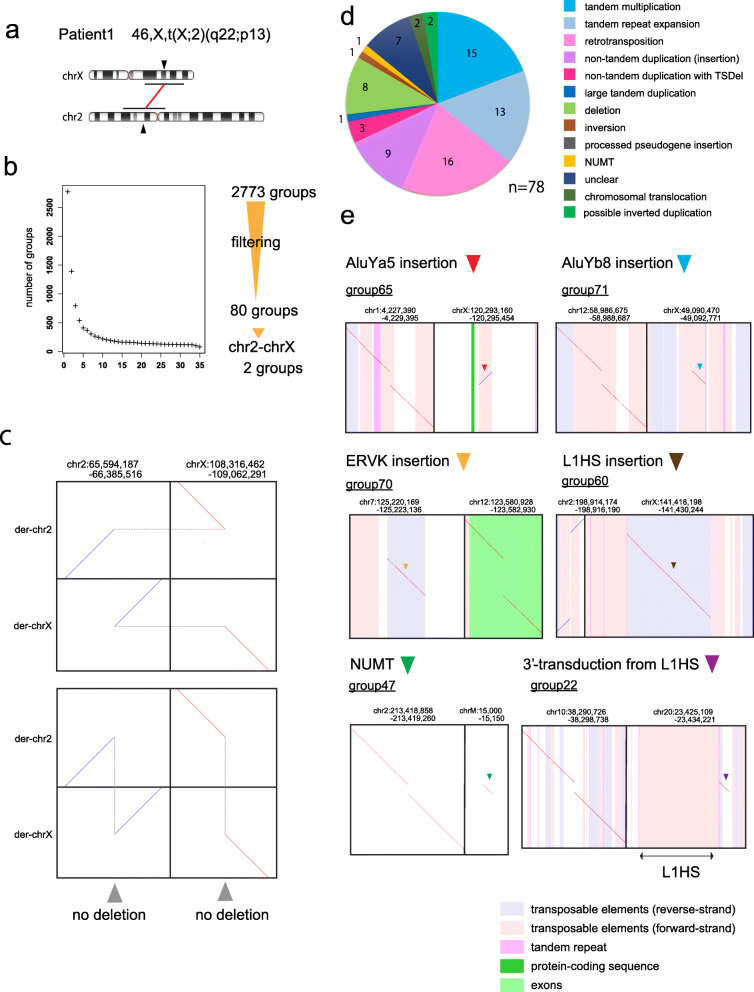
Fig. 4.
Chromosomal rearrangement in patient 1 with 46,X,t(X;2)(q22;p13). a Ideograms showing patient 1’s translocation between chrXq22 and chr2p13. Chromosome images are from NCBI genome decoration (https://www.ncbi.nlm.nih.gov/genome/tools/gdp). b Filtering out rearrangements shared with 33 controls. Finally, 80 groups of reads with patient-only rearrangements are found. Two of the 80 groups show reciprocal chr2-chrX translocation. c Dotplot of reconstructed derivative chromosomes shows reciprocal balanced chromosomal translocation (upper panel: horizontal dotted gray lines join the parts of each derivative chromosome; lower panel: vertical dotted gray lines join fragments that come from adjacent parts of the reference genome, showing there is no large deletion or duplication). d Pie chart of the types of rearrangement. TSDel target site deletion, NUMT nuclear mitochondrial DNA insertion. e Examples of retrotransposition and NUMT insertion (the alignments to retrotransposons, e.g., the AluYa5 in chrX, often have low confidence, indicating uncertainty that this specific AluYa5 is the source)