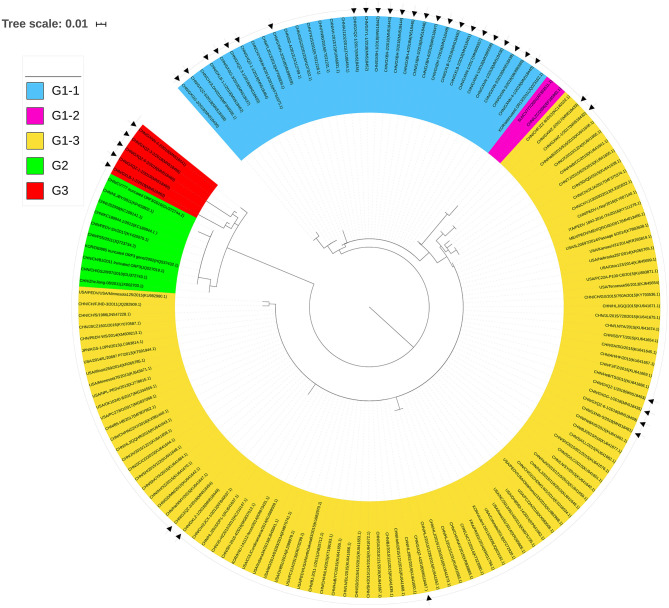
Figure 5.
Phylogenetic tree of ORF3 of PEDV samples based on the amino acid sequences. Sequences of reference and vaccine strains were obtained from GeneBank, the accession numbers are shown in Supplementary Table 1. Tree topology was constructed using the Poisson model and bootstrap re-sampling (1,000 data sets) of the multiple alignments was used to test the statistical robustness of the trees obtained by the neighbor-joining method from MEGA 5.2. Each PEDV strain is indicated in the following format: Country origin (three letter code: CHN, China; FRA, France; ITA, Italy; JPN, Japan; KOR, Korea; MEX, Mexico; SUI, Switzerland; USA, the United States)/strain name/year of sample collection (Genbank accession number). The Group 3 consisted of Guangxi naturally truncated strains which were coded in red. G1-1 subgroup, G1-2 subgroup, G1-3 subgroup, and Group 2 were coded in blue, pink, yellow, and green, respectively. The triangle symbols represent the Guangxi field strains obtained in this study.