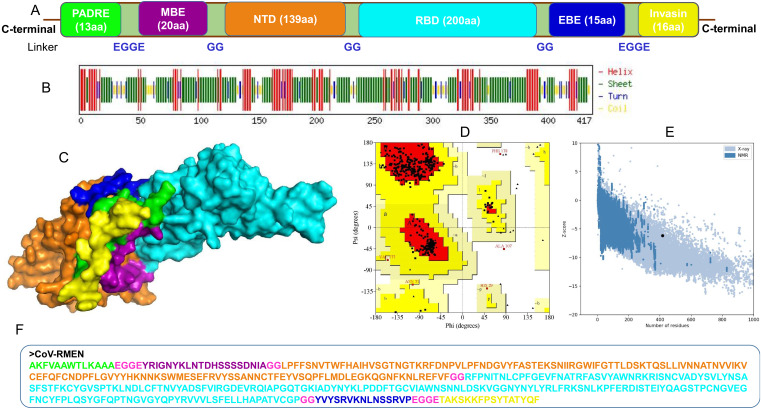
Figure 3. Design, construction and structural validation of multi-epitope vaccine candidate (CoV-RMEN) for SARS-CoV-2.
(A) Structural domains and epitopes rearrangement of CoV-RMEN, (B) secondary structure of CoV-RMEN as analyzed through CFSSP: Chou and Fasman secondary structure prediction server , (C) final tertiary structure of CoV-RMEN (surface view) obtained from homology modelling on Phyre2 in which domains and epitopes are represented in different colors (PADRE-smudge; membrane B-cell epitope, MBE-magenta; N-terminal domain, NTD-orange; receptor-binding domain, RBD-cyan; envelop B-cell epitope, EBE-blue; invasin-yellow), (D) validation of the refined model with Ramachandran plot analysis showing 94.7%, 4.8% and 0.5% of protein residues in favored, allowed, and disallowed (outlier) regions respectively, (e) ProSA-web, giving a Z-score of −6.17, and (f) the finally predicted primary structure of the CoV-RMEN.