Figure 5. An unbiased combinatorial strategy for identifying commensal correlates of protection against and susceptibility to V. cholerae colonization.
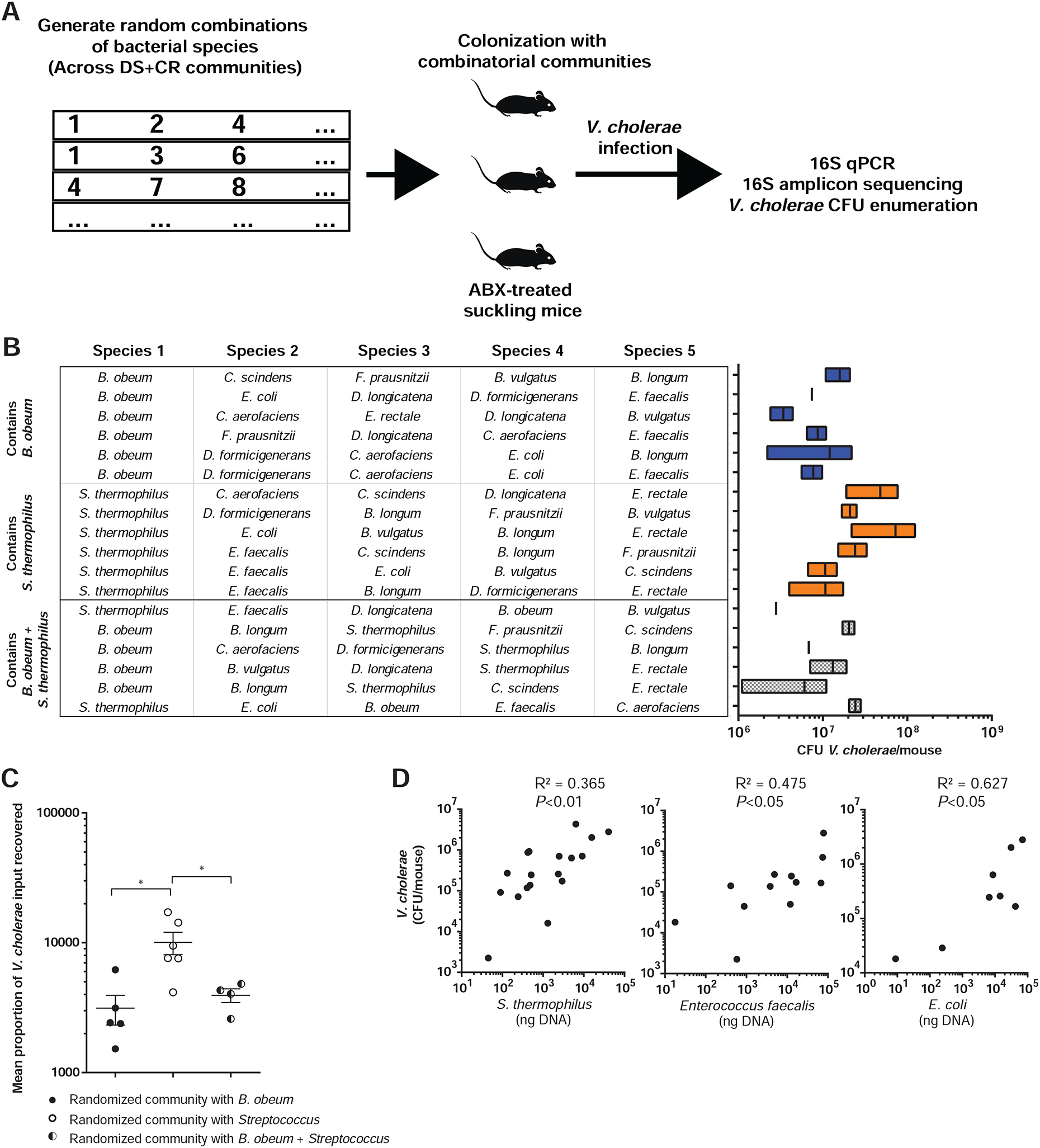
(A) Combinatorial strategy. (B) 5-member microbiome embodiments randomly generated using CR/DS members (left). Resulting V. cholerae infection of antibiotic-treated suckling mice containing defined microbiome embodiments are shown at right. (C) Mean V. cholerae colonization in suckling mice bearing communities containing B. obeum or Streptococcus species alone and in combination. Data normalized across experiments as fold CFU gavaged V. cholerae recovered after infection. (D) Abundance of DS member species in randomized microbiomes correlated to resulting V. cholerae abundance after infection. Points represent mice receiving different microbiome embodiments. * P<0.05, ** P<0.01 (Mann-Whitney U-test); n.s. not significant. Error bars represent mean ± SEM.
