Fig. 10.4.
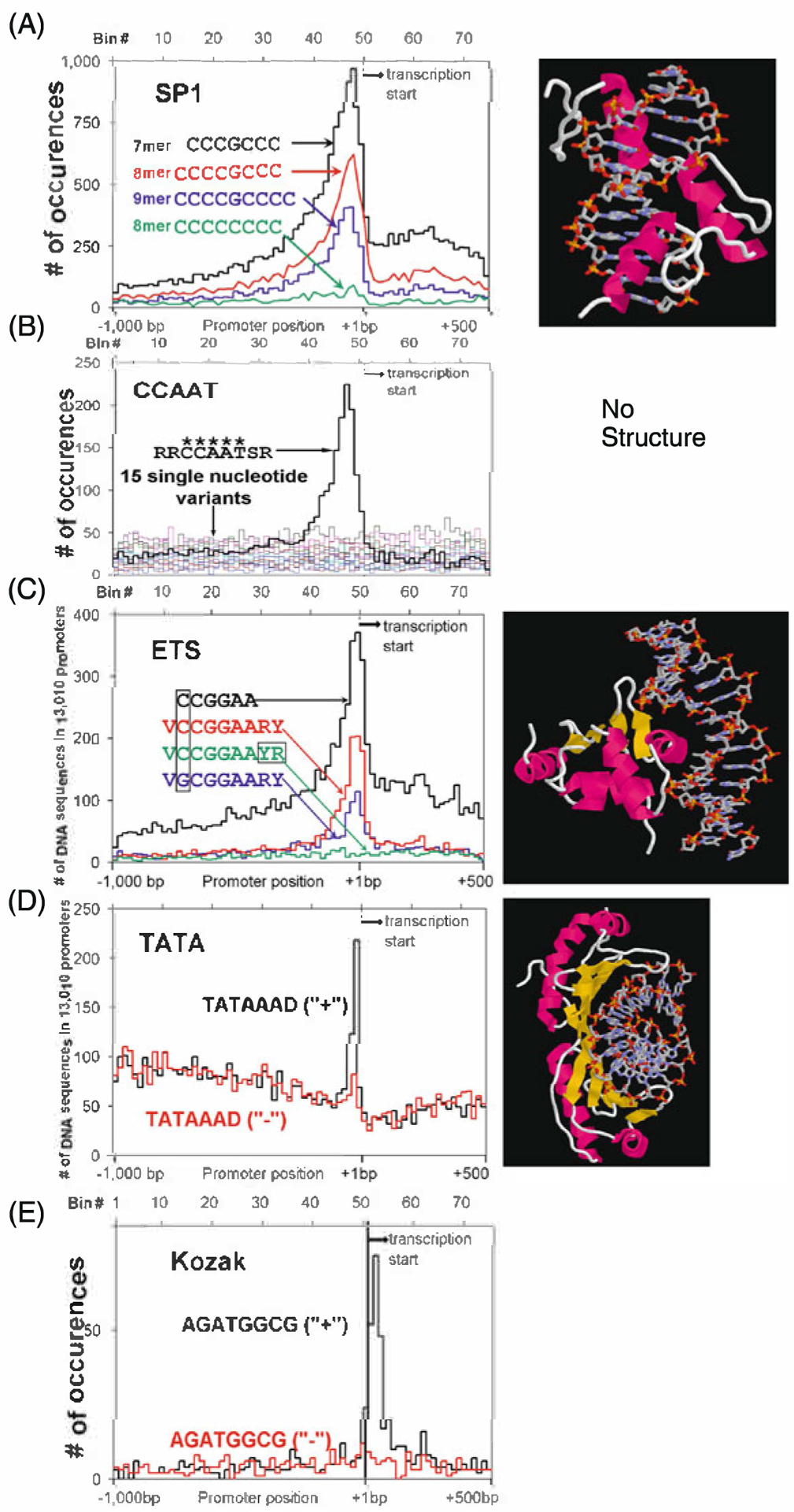
Distribution of non-palindromic TFBS in promoters. We include both the distribution of the TFBS and the X-ray crystal structure if it exists. a SP1 sequences (CCCGCCC, CCCCGCCC, CCCCGCCCC) and a non-peaking single base variation (CCCCCCCC). Crystal structure of a three zinc finger protein bound to DNA. b The CCAAT consensus RRCCAATSR and the 15 single base variants of the central CCAAT. Note the 5-mer CCAAT is needed for there to be any localization in the proximal promoter. No crystal structure is available. c ETS core (CCGGAA), consensus sequence (VCCGGAARY), and a peaking (VGCGGAARY) and non-peaking VCCGGAAYR variant. Crystal structure of ETS bound to DNA d Strand specific localization of the TATAAAD sequence. Note both the high background and the sharpness of the peak. Crystal structure of TATA bound to DNA. This is the only protein DNA complex presented here without an α-helix in the major groove of DNA. e Kozak sequence (AGATGGCG) on the plus strand (+) and minus strand (−). Again, note the DNA strand dependence of the localization of this sequence
