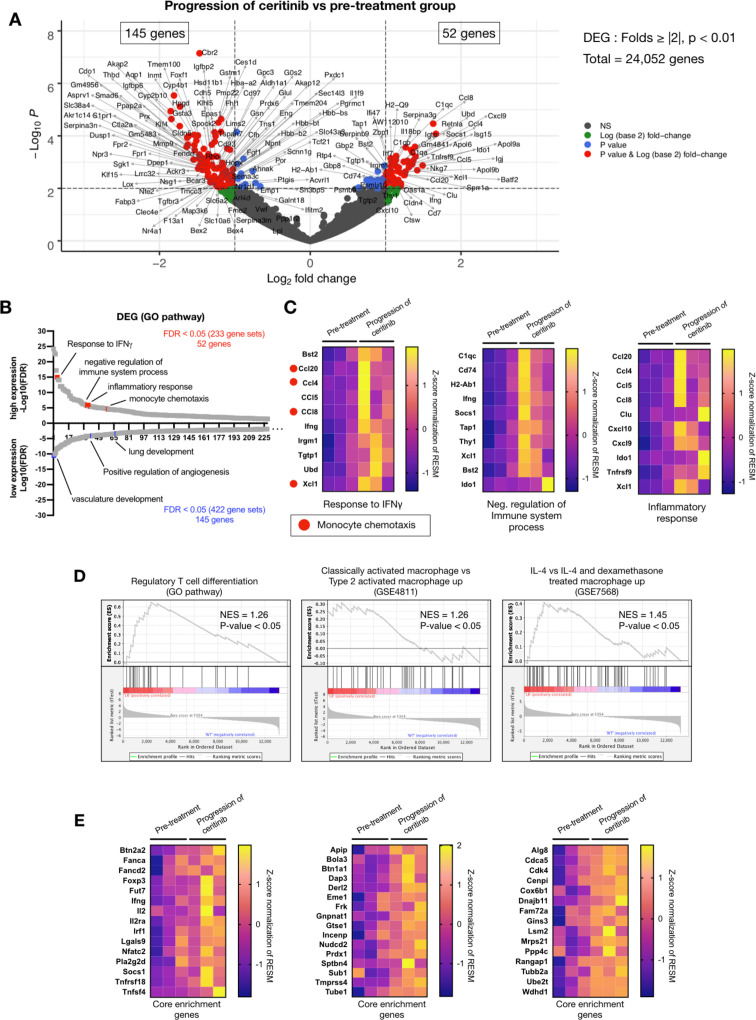
Figure 6.
Differentially expressed genes (DEGs) and gene set enrichment analysis (GSEA) between progression of ceritinib and pretreatment groups. (A) A total of 24 052 genes are presented in Volcano plots to compare differentially expressed genes in postceritinib resistance and pretreatment tumors. DESeq2 package was used for folds of gene expression and statistical analysis (folds ≧|2|, p<0.01). The individual gene expression and p values were displayed as colored dots (gray=no-significant and no-folds changes, green=folds >|2|, blue ≤0.01, red=folds ≧|2| and p<0.01). Lower expression of 145 genes and higher expression of 52 genes were identified in postceritinib resistance tumor. (B) The selected DEGs were analyzed by the gene set enrichment with higher expressing 52 genes and lower expressing 145 genes. GO pathway was applied to analyze the meaning of previous results. The presented gene sets were significantly enriched gene sets in GO pathway (FDR >0.05). The indicated gene sets were highly related to previous results (red square dots=high expressed genes, blue square dots=low expressed genes). (C) The heatmaps were presented to show core enrichment genes of previously related gene sets; IFN-γ, negative regulation of immune system process and inflammatory response. The gradient key color presents the expression of individual genes (Z-score normalized). (D) GSEA with MSigdb.v7.0. (12,999 gene sets). Among the significant gene sets, regulatory T cell differentiation (GO), classically activated macrophage versus type 2 activated macrophage up (GSE4811), and IL-4 versus IL-4 and dexamethasone-treated macrophage up (GSE7568) were demonstrated with normalized enrichment score (NES)and p value. (E) The heatmaps were presented to show core enrichment genes.