Abstract
Introduction
The molecular pathogenesis of liver cancer remains unclear; some ncRNAs have been considered as potential drug targets for cancer treatment. LncRNA PSMG3‑AS1 has been reported to promote breast cancer, while its role in hepatocellular carcinoma (HCC) is unknown.
Methods
Bioinformatics analysis was conducted to investigate the relationship between miR-143-3p and PSMG3-AS1. RT-qPCR was used to detect the expression levels of miR-143-3p and PSMG3-AS1 and the correlation between them in HCC. The survival curve was used to analyze the effect of PSMG3-AS1 on the prognosis of liver cancer. RT-qPCR was used to detect the effect of different concentration gradients of miR-143-3p on PSMG3-AS1. CCK8 and clone formation experiments were used to examine the role of miR-143-3p and PSMG3-AS1 in regulating the proliferation of SNU-182 and SNU-398 cells.
Results
Our preliminary bioinformatics analysis showed that miR-143-3p can target PSMG3-AS1. We, therefore, analyzed the interaction between PSMG3-AS1 and miR-143-3p in HCC. We found that PSMG3-AS1 was upregulated, while miR-143-3p was downregulated in HCC. The expression levels of PSMG3‑AS1 and miR-143-3p were closely and inversely correlated with each other. High expression levels of PSMG3‑AS1 predicted poor survival. In HCC cells, overexpression of PSMG3-AS1 led to increased proliferation rates. Overexpression of miR-143-3p played an opposite role and reversed the effect of overexpression of PSMG3‑AS1.
Discussions
miR-143-3p may target PSMG3‑AS1 to inhibit the proliferation of HCC cells.
Keywords: hepatocellular carcinoma, lncRNA PSMG3-AS1, miR-143-3p, proliferation
Introduction
As a type of malignancy contains intrahepatic cholangiocarcinoma (ICC) and hepatocellular carcinoma (HCC), liver cancer is a major burden on the public health.1 Liver cancer in 2018 alone affected 841,080 new cases (4.7% of all new cancer cases), which ranked the 7th most common type of malignancies.2 In contrast, due to its extreme aggressive nature, liver cancer caused 781,631 deaths (8.2% of all cancer deaths), which made it the 3rd most common cause of cancer-related deaths.2 Liver cancer is mainly caused by the use of tobacco and alcohol as well as the infections of hepatitis B virus (HBV) and C virus (HCV).3,4 Although our understanding of the mechanism of HBV and HCV infections has been significantly increased during the past decade,5,6 the molecular pathogenesis of liver cancer remains unclear.
The development and progression of liver cancer involves multiple signaling pathways.7,8 Functional analyses of the signaling pathways involved in this disease provide novel insights into the development of targeted therapies for cancer treatment.7,8 Although non-coding RNAs (ncRNAs), such as microRNAs (miRNAs) and long (> 200 nt) ncRNAs (lncRNAs), are not involved in protein coding, regulate the expression of cancer-related genes to promote or suppress cancer development.9,10 In effect, some ncRNAs have been considered as potential drug targets for cancer treatment.11 However, the functions of most ncRNAs in cancer biology remain unclear. PSMG3‑AS1 is a novel tumor-suppressive lncRNA characterized in breast cancer,12 while its role in HCC, a major subtype of liver, remains unknown. Our preliminary bioinformatics analysis showed that PSMG3‑AS1 may be targeted by miR-142-3p, which is a well-characterized tumor suppressor.13 This study was therefore carried out to investigate the interaction between PSMG3-AS1 and miR-142-3p in HCC.
Patients and Methods
Patients, Treatment and Follow-Up
A total of 66 HCC patients (40 males and 26 females, 43 to 69 years old, mean age 55.1 ± 6.3 years old) admitted at the First Affiliated Hospital of Anhui Medical University between May 2012 and May 2014 were enrolled. All HCC patients were new cases diagnosed by histopathological exams (Figure S1). No therapy was initiated, and patients with other clinical disorders were excluded. According to their medical record, 20 cases were HBV positive only, 17 cases were HCV positive only, 13 cases were positive for both HBV and HCV, and 16 cases were negative for both HBV and HCV. Treatments were determined based on their AJCC stages. The 68 patients included 9 stage I, 10 stage II, 24 stage III and 23 cases stage IV cases. Surgical resection, chemotherapy, radiotherapy, targeted therapy or combined therapies were performed. All patients signed the written informed consent. All patients were followed up for 5 years after admission. All patients completed the 5-year follow-up. Their survival conditions were recorded and used for survival analysis. This study was approved by the Ethics Community of the aforementioned hospital and was conducted in accordance with the Declaration of Helsinki.
Tissues and HCC Cells
All patients were diagnosed by histopathological biopsy. During biopsy, both HCC and adjacent non-tumor tissues (within 3 cm around tumors) were collected from each patient. All tissue samples were tested by histopathological analysis and were stored in liquid nitrogen before use. SNU-182 and SNU-398 cells obtained from ATCC (USA) were used to perform in vitro experiments. Cell culture medium was composed of 10% FBS and 90% RPMI 1640 medium. Cells were cultivated in a 5% CO2 incubator at 37 °C with 95% humidity.
RNA Preparations and RT-qPCR
Total RNAs were isolated from biopsies and cultivated cells using RNAzol (Sigma-Aldrich). All RNA samples were digested with DNase I (Sigma-Aldrich) to remove genomic DNAs. RNA concentrations were measured by NanoDrop 2000 Spectrophotometer (Thermo Scientific). RNA samples with satisfactory qualities were subjected to reverse transcriptions using QuantiTect Reverse Transcription Kit (QIAGEN) to synthesize cDNA samples. With cDNA samples as template, qPCR reactions were prepared using SYBR® Green Quantitative RT-qPCR Kit (Sigma-Aldrich). The expression levels of PSMG3-AS1 were measured with 18S rRNA as endogenous control. The expression levels of mature miR-143-3p were measured using All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia). U6 was used as the endogenous control of miR-143-3p. PCR reactions were performed in triplicate. Expression levels were normalized using 2−ΔΔCt method. Primer sequences were as following: 5ʹ-GAAGCAGAACCAACGCACAG-3ʹ (forward) and 5ʹ-GCATAATCCAATCCCTCAAGAA-3ʹ (reverse) for PSMG3-AS1; 5ʹ-CTGGCGTTGAGATGAAGCAC-3ʹ (forward) and reverse, 5ʹ-CAGAGCAGGGTCCGAGGTA-3ʹ (reverse) for miR-143-3p.
Cell Transfection
Expression vector of PSMG3‑AS1 was constructed using pcDNA 3.1 (Sigma-Aldrich) as the backbone. miR-143-3p mimic and negative control (NC) miRNA were purchased from Sigma-Aldrich. SNU-182 and SNU-398 cells were collected at about 80% confluence and counted, followed by transfecting 10 nM vector or 10~100 nM siRNA into 106 cells using lipofectamine 2000 (Invitrogen). Control (C) cells in all transfections were cells without transfections. NC cells were empty vector- or NC miRNA-transfected cells. Subsequent experiments were performed 48 h post-transfection.
Cell Proliferation Assay
Cell Counting Kit-8 kit (CCK-8, Dojindo) was used to perform cell proliferation assays on SNU-182 and SNU-398 cells collected at 48 h post-transfection. In brief, 3000 cells were transferred to each well of a 96-well cell culture plate. Cells were cultivated under aforementioned conditions, and cell collections were performed every 24 h for a total of 4 days. At 4 h before cell collection, CCK-8 solution was added into each well to reach 10% final concentration. After cell collection, OD values were measured at 450 nm.
Colony Formation Assay
For colony formation assay, the transfected cells were seeded into 6-well plates (4000 cells/well) and incubated in DMEM with 10% FBS. After 2 weeks, cells were fixed by methanol and stained by 0.1% crystal violet. The colony numbers were then counted.
Statistical Analysis
Three biological replicates were included in each experiment. Mean ± SEM values were used to express all data. Paired t-test was used to compare non-tumor and HCC tissues. ANOVA (one-way) and Tukey’s test were used to compare differences among multiple groups. Spearman’s rank correlation coefficient was used for correlation analysis. Survival analysis was performed by dividing the 66 HCC patients into high and low PSMG3‑AS1 level groups (n = 33). Survival curves were plotted using GraphPad Prism 6 software and compared by Log-rank test. P < 0.05 was statistically significant.
Results
The Expression of PSMG3-AS1 and miR-143-3p Was Altered in HCC
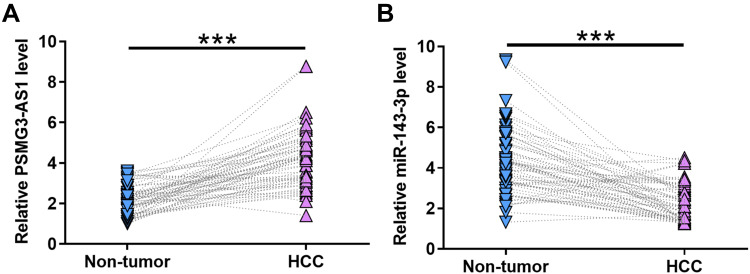
The expression levels of PSMG3-AS1 and miR-143-3p in both HCC and non-tumor tissues were measured by RT-qPCR assays. Compared to non-tumor tissues, the expression levels of PSMG3‑AS1 were significantly higher in HCC tissues (Figure 1A, p < 0.001). In contrast, the expression levels of miR-143-3p were significantly lower in HCC tissues than that in non-tumor tissues (Figure 1B, p < 0.001). It is worth noting that the expression levels of PSMG3-AS1 and miR-143-3p were not significantly different among different HBV and HCV infection groups.
Figure 1.
The expression of PSMG3-AS1and miR-143-3p were altered in HCC. The expression levels of PSMG3-AS1 (A) and miR-143-3p (B) in both HCC and non-tumor tissues were measured by performing RT-qPCR assays. PCR reactions were performed in 3 replicates and mean values were presented and compared. ***p < 0.05.
The Expression Levels of PSMG3-AS1 and miR-143-3p Were Closely and Inversely Correlated
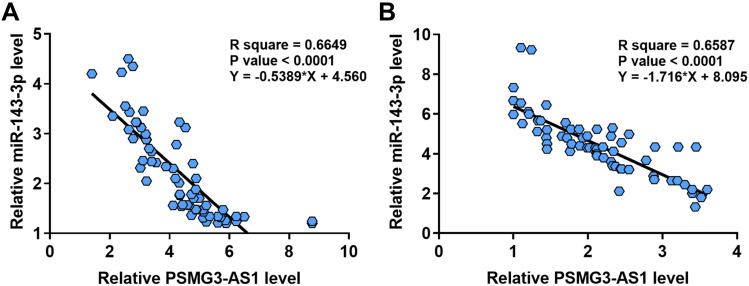
To find out whether PSMG3-AS1 and miR-143-3p were only related to tumorigenesis, Spearman’s rank correlation coefficient was performed to analyze the correlation between the expression levels of PSMG3-AS1 and miR-143-3p across both HCC and non-tumor tissues. It was observed that the expressions of PSMG3-AS1 and miR-143-3p were inversely and significantly correlated with each other across both HCC (Figure 2A) and non-tumor (Figure 2B) tissues.
Figure 2.
The expression levels of PSMG3-AS1 and miR-143-3p were closely and inversely correlated. Spearman’s rank correlation coefficient was performed to analyze the correlation between expression levels of PSMG3-AS1 and miR-143-3p across both HCC (A) and non-tumor tissues (B).
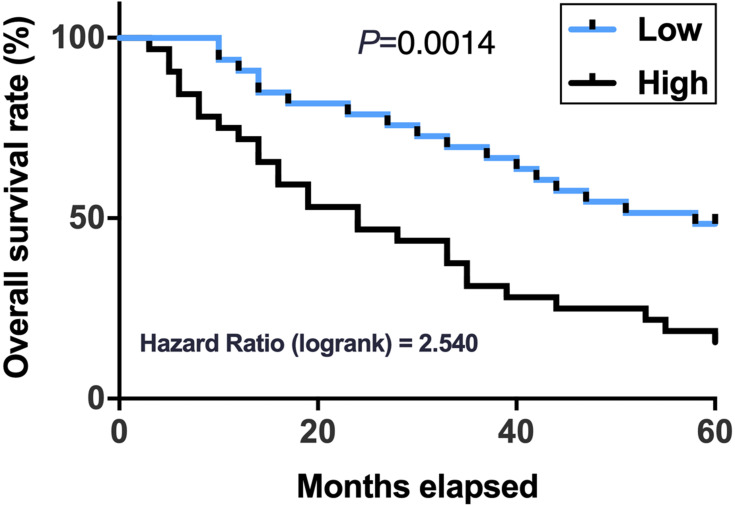
High Expression Levels of PSMG3-AS1 in HCC Predicted Poor Survival
Survival curve analysis was performed to evaluate the prognostic value of PSMG3-AS1 for HCC. Compared to low PSMG3-AS1 level group, patients in high PSMG3-AS1 level group experienced a significantly lower overall survival rate (Figure 3).
Figure 3.
High expression levels of PSMG3-AS1 in HCC predicted poor survival. Survival analysis was performed by dividing the 66 HCC patients into high and low PSMG3-AS1 level groups (n = 33). Survival curves were plotted using GraphPad Prism 6 software and compared by Log-rank test.
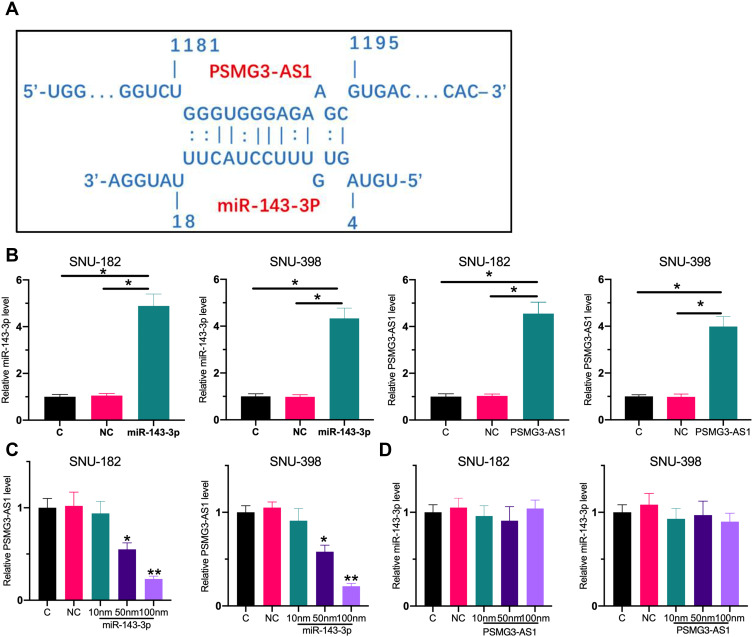
miR-143-3p May Downregulate PSMG3-AS1 by Direct Targeting
The interaction between miR-143-3p and PSMG3-AS1 was predicted by IntaRNA.14 It was observed that miR-143-3p might target PSMG3-AS1 (Figure 4A). To confirm this, SNU-182 and SNU-398 cells were transfected with miR-143-3p mimic or PSMG3-AS1 expression vector, followed by the confirmation of the overexpression of miR-143-3p and PSMG3-AS1 by RT-qPCR (Figure 4B, p < 0.05). Compared to C and NC groups, overexpression of miR-143-3p led to significantly downregulated PSMG3-AS1 (Figure 4C, p < 0.05). In contrast, overexpression of PSMG3-AS1 did not affect the expression of miR-143-3p (Figure 4D).
Figure 4.
MiR-143-3p may downregulate PSMG3-AS1 by direct targeting. (A) The IntaRNA tool was used to predict the interaction between the retrieved miR-143-3p and PSMG3-AS1 sequence. The results show that there is a 12 bp sequence complementarity between PSMG3‑AS1 and miR-143-3p. This indicates that there is a possibility of binding between PSMG3-AS1 and miR-143-3p. SNU-182 and SNU-398 cells were transfected with miR-143-3p mimic or PSMG3-AS1 expression vector, followed by the confirmation of the overexpression of miR-143-3p and PSMG3-AS1 by RT-qPCR (B). The effects of overexpression of miR-143-3p on PSMG3-AS1 (C) and the effects of overexpression of PSMG3-AS1 on miR-143-3p (D) were also analyzed by RT-qPCR. All experiments were performed in triplicate and mean values were compared. *p < 0.05; **p < 0.01.
miR-143-3p Downregulates PSMG3-AS1 to Inhibit Cell Proliferation
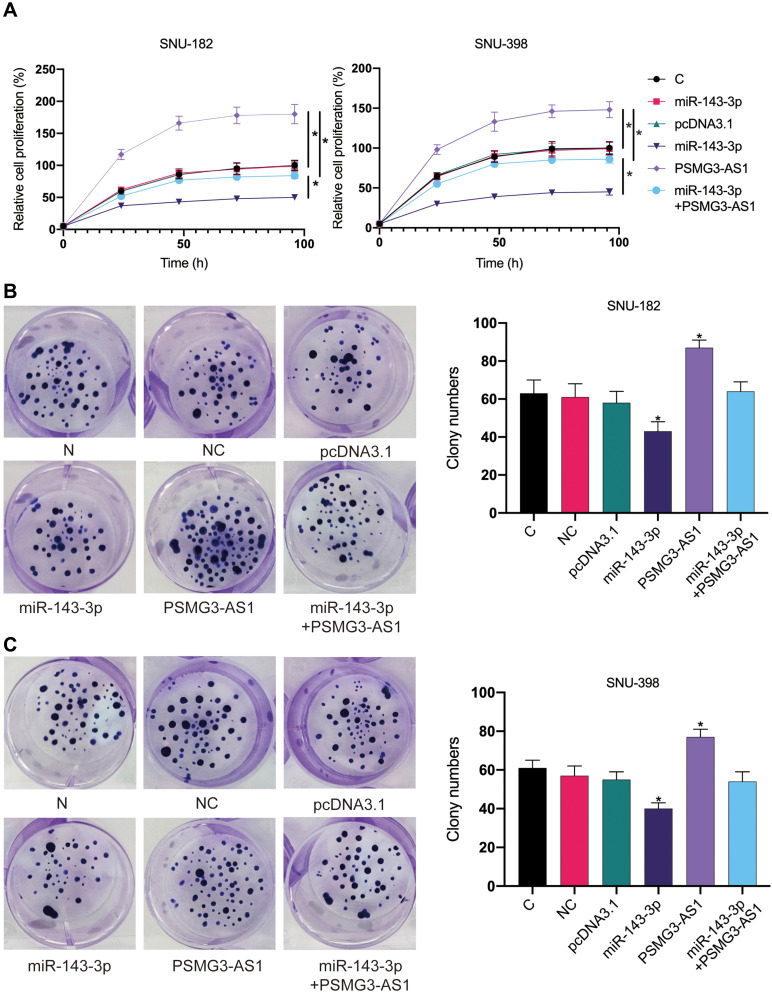
CCK-8 analysis (Figure 5A) and clone formation experiments (Figure 5B and C) were conducted to explore the role of miR-143-3p and PSMG3-AS1 in regulating the proliferation of SNU-182 and SNU-398 cells. The cell proliferation rate of the PSMG3-AS1 group increased significantly, while the miR-143-3p group decreased. Compared with group C, there was no significant difference in cell proliferation between PSMG3-AS1 and miR-143-3p cotransfection groups (Figure 5, p <0.05). This shows that miR-143-3p and PSMG3-AS1 play the opposite role and reduce the effect of PSMG3-AS1 overexpression on cell proliferation.
Figure 5.
MiR-143-3p downregulates PSMG3-AS1 to inhibit cell proliferation. (A). CCK-8 assay was performed to analyze the roles of miR-143-3p and PSMG3-AS1 in regulating the proliferation of SNU-182 and SNU-398 cells. The cell proliferation rate in the PSMG3-AS1 group increased significantly, while the miR-143-3p group decreased. There was no significant difference in cell proliferation between the PSMG3-AS1 and miR-143-3p co-transfection groups compared with the group C. B and C are colony formation experiments in SNU-182 (B) and SNU-398 (C). The colony formation of miR-143-3p group was significantly lower than that of group C, while the PSMG3-AS1 group was higher. The number of colonies formed in PSMG3-AS1 and miR-143-3p groups was not significantly different from that in group C. All experiments were performed in 3 replicates and mean values were presented and compared. *p < 0.05.
Discussion
Increasing evidence has shown that long non-coding RNAs (lncRNAs) play important roles in tumor progression. In this study, we analyzed the interaction between miR-143-3p and PSMG3-AS1 in HCC. We found that miR-143-3p and PSMG3-AS1 were both dysregulated in liver cancer and miR-143-3p may target PSMG3-AS1 to suppress HCC cell proliferation.
The functionality of PSMG3-AS1 has only been explored in breast cancer.12 It was reported that PSMG3-AS1 was downregulated in breast cancer and may sponge miR-143-3p to promote the migration and proliferation of cancer cells.12 This study is the first to report the upregulation of PSMG3-AS1 in HCC. Moreover, overexpression of PSMG3-AS1 in HCC cells led to decreased proliferation of HCC cells. Therefore, our study proved that PSMG3-AS1 was also likely an oncogenic lncRNA in HCC.
More than 50% of HCC patients diagnosed at an early stage can live longer than 5 years after the initial diagnosis.15 However, most HCC patients are diagnosed at advanced stages, which lack radical treatment. As a consequence, the survival rate of most HCC patients remains low. In this study, we proved that high expression levels of PSMG3-AS1 were closely correlated with poor survival of HCC patients. Therefore, measuring the expression levels of PSMG3-AS1 before treatment may be used to predict the survival of HCC patients and guide the selection of therapeutic approaches. The analysis of the lncRNA co-expression module of ovarian cancer found that 16 lncRNAs including PSMG3-AS1 were significantly associated with the pathological diagnosis, lymphatic vessel infiltration, tissue source site and the age of patients with vascular infiltration.16 PSMG3-AS1 may be related to the prognosis of ovarian cancer. However, the prognostic accuracy remains to be further tested.
miR-142-3p has been proven as a tumor-suppressive miRNA in multiple types of malignancy.13,17 In osteosarcoma, miR-143-3p can inhibit the proliferation, migration and invasion of osteosarcoma by targeting FOSL2.18 In breast cancer, miR-143-3p can inhibit the proliferation, cell migration and invasion of human breast cancer cells by regulating the expression of MAPK7.19 A recent study reported that miR-142-3p is downregulated in HCC and its overexpression can inhibit cell proliferation in HCC.17 Our study confirmed its downregulation in HCC and its inhibitory role in regulating HCC cell proliferation. We proved that, besides protein coding genes, such as Bach-1 and LDHA,13,17 miR-142-3p can also target an lncRNA, which is PSMG3-AS1, to suppress cancer development. However, other mechanisms may also exist, and more studies are still needed.
Conclusion
In conclusion, PSMG-AS1 is overexpressed in HCC. miR-142-3p can target PSMG3-AS1 to suppress cancer cell proliferation.
Funding Statement
We thank the financial support from Scientific Research Fund of Anhui Medical University (NO. 2015xkj089).
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Seehawer M, Heinzmann F, D’Artista L, et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature. 2018;562(7725):69–75. doi: 10.1038/s41586-018-0519-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Petrick JL, Campbell PT, Koshiol J, et al. Tobacco, alcohol use and risk of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: the Liver Cancer Pooling Project. Br J Cancer. 2018;118(7):1005–1012. doi: 10.1038/s41416-018-0007-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J Cancer. 2018;142(12):2471–2477. doi: 10.1002/ijc.31280 [DOI] [PubMed] [Google Scholar]
- 5.Negash AA, Olson RM, Griffin S, Gale M Jr. Modulation of calcium signaling pathway by hepatitis C virus core protein stimulates NLRP3 inflammasome activation. PLoS Pathog. 2019;15(2):e1007593. doi: 10.1371/journal.ppat.1007593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang KM, Traum D, Park JJ, et al. Distinct phenotype and function of circulating Vdelta1+ and Vdelta2+ gammadeltaT-cells in acute and chronic hepatitis B. PLoS Pathog. 2019;15(4):e1007715. doi: 10.1371/journal.ppat.1007715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanasekaran R, Bandoh S, Roberts LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000 Res. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152(4):745–761. doi: 10.1053/j.gastro.2016.11.048 [DOI] [PubMed] [Google Scholar]
- 9.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi: 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi: 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Y, Fan Y, Zhao G, et al. Novel lncRNA PSMG3AS1 functions as a miR1433p sponge to increase the proliferation and migration of breast cancer cells. Oncol Rep. 2019. doi: 10.3892/or.2019.7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansoori B, Mohammadi A, Ghasabi M, et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J Cell Physiol. 2019;234(6):9816–9825. doi: 10.1002/jcp.27670 [DOI] [PubMed] [Google Scholar]
- 14.Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45(W1):W435–W439. doi: 10.1093/nar/gkx279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Momin BR, Pinheiro PS, Carreira H, Li C, Weir HK. Liver cancer survival in the United States by race and stage (20012009): findings from the CONCORD-2 study. Cancer. 2017;123(Suppl 24):5059–5078. doi: 10.1002/cncr.30820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Zhan X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J. 2019;10(3):273–290. doi: 10.1007/s13167-019-00175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua S, Liu C, Liu L, Wu D. miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochem Biophys Res Commun. 2018;496(3):947–954. doi: 10.1016/j.bbrc.2018.01.112 [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Dai G, Yu L, Hu Q, Chen J, Guo W. miR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci Rep. 2018;8(1):606. doi: 10.1038/s41598-017-18739-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian Y, Teng Y, Li Y, et al. MiR-143-3p suppresses the progression of nasal squamous cell carcinoma by targeting Bcl-2 and IGF1R. Biochem Biophys Res Commun. 2019;518(3):492–499. doi: 10.1016/j.bbrc.2019.08.075 [DOI] [PubMed] [Google Scholar]