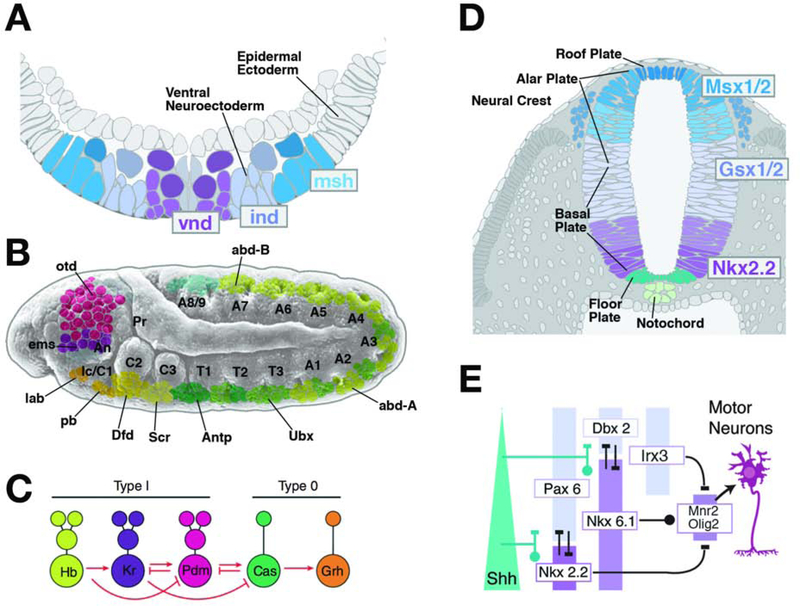
Figure 3.
Neural proliferation and specification of neural fate. (A, B) Transcriptional regulators determining neural fate subdivide the neuroectoderm into discrete domains. (A) depicts schematic cross section of Drosophila ventral neuroectoderm following neuroblast delamination; the genes vnd, ind and msh are expressed in longitudinal columns of neuroectoderm and resulting neuroblasts. (B) shows expression pattern of Hox genes (lab, pb, Dfd, Scr, Antp, Ubx, abdA, abdB) and head gap genes (otd, ems) in discrete domains of neuroectoderm and neuroblast layer along the antero-posterior axis. (C) A cassette of sequentially expressed transcription factors, conisting of Hunchback (Hb), Kruppel (Kr), Nubbin (Nub), Castor (Cas) and Grainyhead (Grh), specifies the fate of embryonic Drosophila neuroblasts. Direct interactions between these factors orchestrate the temporal dynamics of their expression (from Allan and Thor, 2015, with permission). (D, E) The expression pattern of transcriptional regulators, many of them homologous to those discovered in Drosophila (see conserved color coding between panels A and D), divide the neural tube into longitudinal columns. The vnd homolog Nkx2.2, as well as Nkx6.1, activated by high levels of the signal Sonic hedgehog (Shh), are expressed in nested columns adjacent to the floor plate (E). Other transcription factors (e.g., Pax6, Dbx2, Irx3, Msx1/2, Gsx1/2) are activated by a dorsally originating BMP signal. Inhibitory interactions among the transcription factors delineate subdomains, as exemplified by the MN domain that gives rise to motor neurons and oligodendrocytes (E).