Fig. 2.
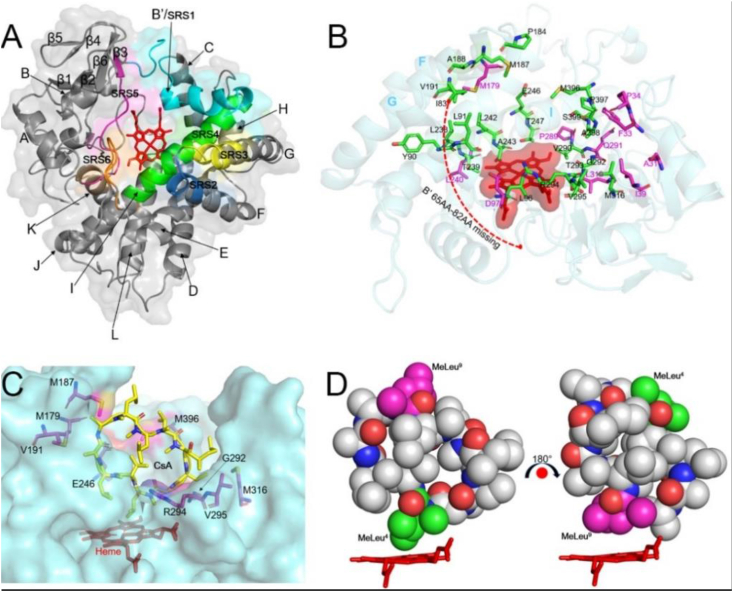
A. Overall structure of CYP-sb21. The conserved secondary structure components (α-helices and β-sheets) are labelled; and the regions of six substrate recognition sites (SRSs) are highlighted in cyan (SRS1, 59–100 aa), blue (SRS2, 178–184 aa), yellow (SRS3, 185–192 aa), green (SRS4, 230–250 aa), magenta (SRS5, 287–297 aa) and orange (SRS6, 395–402 aa). B. The substrate binding pocket for Autodock and MD analyses. The amino acids shown as sticks in green represent the key residues analyzed by both Autodock and MD. The sticks in magenta denote the key amino acids only analyzed by MD. The central heme prosthetic group is shown as stick and surface in red. The unstructured B′ region (56–82 aa) is indicated by a red dashed line. The key helices are labelled by blue capital letters. C. The selected mutation sites. The amino acids that were subject to mutagenesis analysis are shown as sticks in magenta. D. The two productive binding modes of CsA as spheres. MeLeu4 and MeLeu9 are colored in green and magenta, respectively.