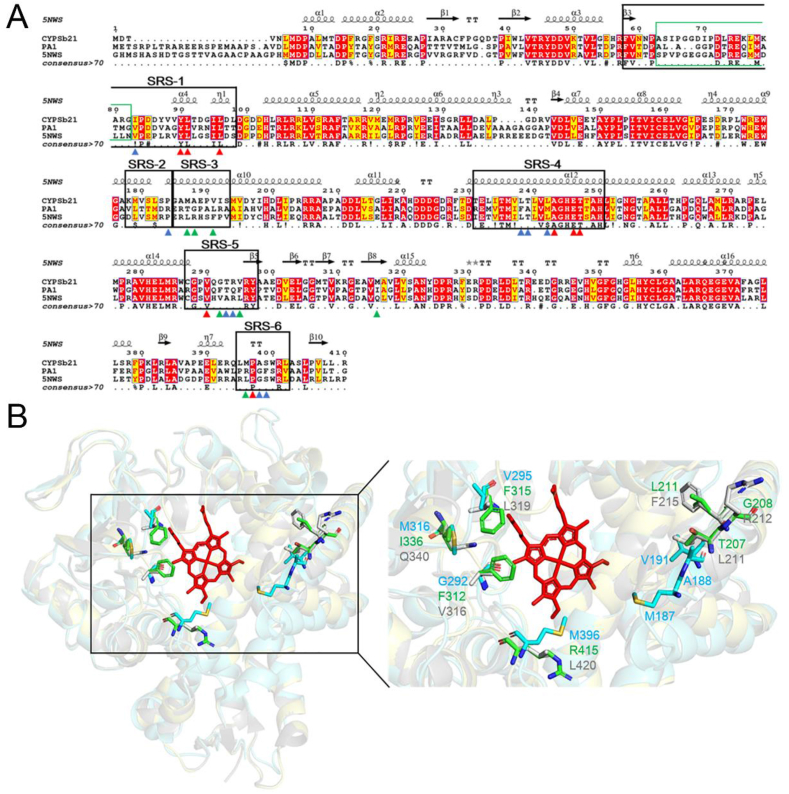
Fig. 4.
Comparison of CYP-sb21, CYP-pa1 and saAcmM (CYP-X). A. Protein sequence alignment of CYP-sb21, CYP-pa1, and saAcmM. The 24 key amino acids in the CsA binding pocket (as described in Fig. 1B) indicated by red, blue, and green triangles denote all identical, two out of three identical, and all different amino acids among the three P450 enzymes, respectively. The regions of six substrate recognition sites (SRSs) are marked by black rectangles. The green rectangle represents the missing region both in CYP-sb21 and saAcmM three-dimensional structures. Sequence analysis was performed using Expresso through the T-COFFEE online service, and the figure was prepared by ESPript 3.0. The η symbol represents a 310-helix. α-Helices and β-strands are indicated as helices and black arrows, respectively; strict β-turns as TT letters and strict α-turns as TTT. B. The 3D structures of CYP-sb21 (PDB ID: 6M4S), CYP-pa1 (simulated by SWISS-MODEL), and saAcmM (PDB ID: 5NWS) shown as cartoon in aquamarine, yelloworange, and gray, respectively. The 7 non-conservative residues among the three P450 enzymes are shown as sticks in aquamarine (CYP-sb21), green (CYP-pa1), and gray (saAcmM).