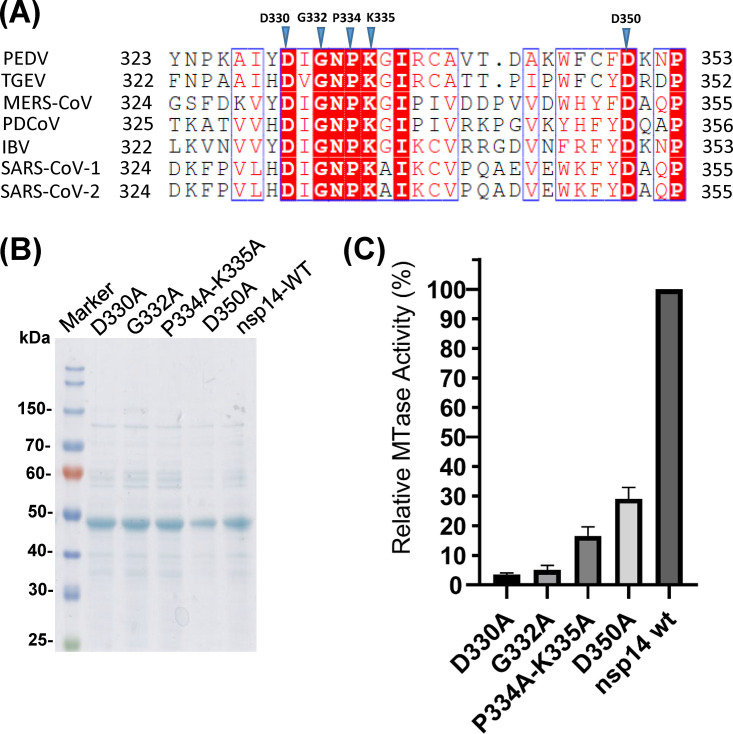
FIG 5.
Mutations to the SAM binding site abolish G-N-7 MTase. (A) Sequence alignment of CoV nsp14 proteins. The G-N-7 MTase regions of PEDV nsp14 (gi: 557844763) and its homologs from Alphacoronavirus genus TGEV (gi: 110746821), Betacoronavirus genus SARS-CoV-1 (gi: 40795428) and SARS-CoV-2 (gi: 1806553187), Deltacoronavirus genus PDCoV (gi: 668361756), and Gammacoronavirus genus IBV (gi: 9626535) were chosen for alignment by ClustalW2. Identical residues are shown in boxes with a solid red background, and conserved residues are highlighted in red. Amino acid residues in the SAM binding residue chosen for mutagenesis are marked with blue triangles. (B) SDS-PAGE analysis of PEDV nsp14 mutants. 6×Hig-tagged PEDV nsp14 mutants were expressed and purified from E. coli. Protein marker is indicated on the left. (C) Analysis of G-N-7 MTase activity of nsp14 mutants. One microgram of PEDV nsp14 protein was incubated with a 296-nt RNA substrate at 37°C for 4 h in the presence of 3H-SAM. The incorporation of 3H-SAM into the RNA substrate was measured by a scintillation counter. Data are the averages from three independent experiments ± standard deviations.