FIG 5.
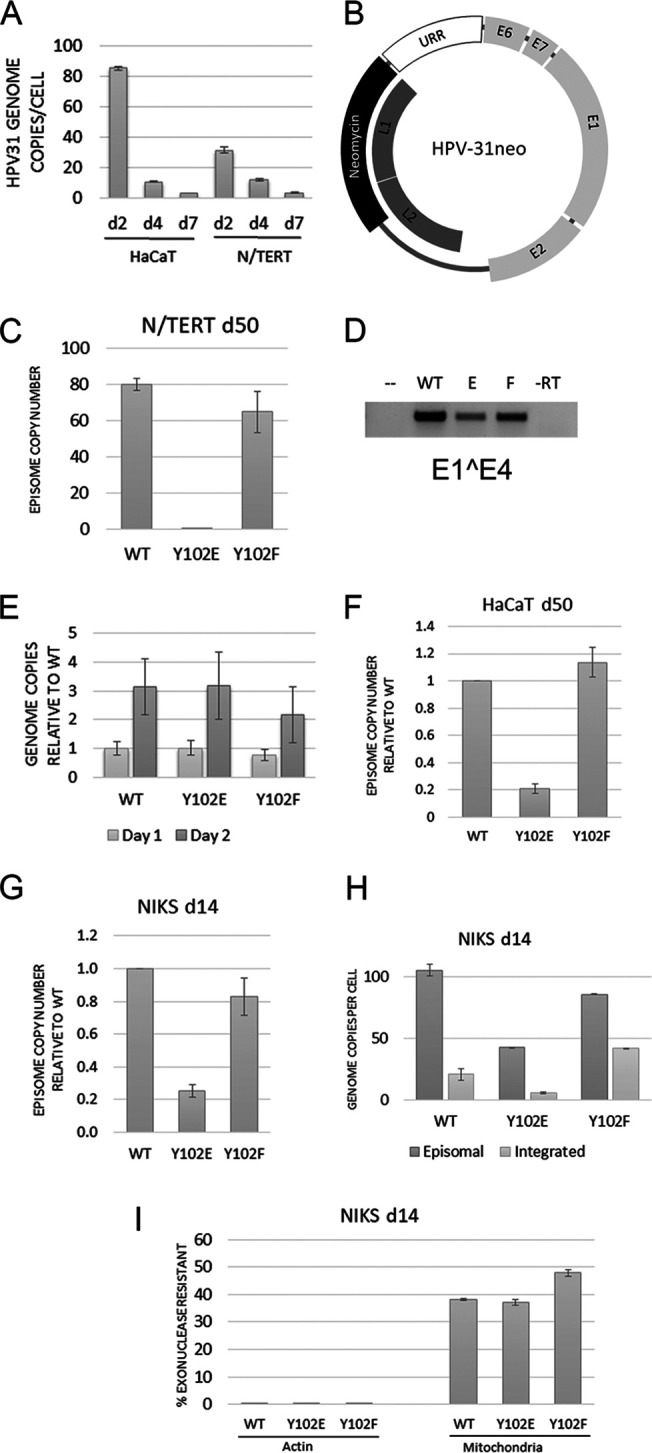
HPV-31 E2 Y102E prevents episomal maintenance. (A) HaCaT and N/TERT cells infected with HPV-31 quasivirus were harvested at days 2, 4, and 7 postinfection. Total DNA was isolated and amplified with primers to the long control region (LCR). (B) Schematic of HPV-31neo genome. The inserted neomycin cassette replaced parts of the L1 and L2 genes in the HPV-31 genome. (C) N/TERT cells transfected with WT, Y102E, or Y102F HPV-31neo genomes were stably maintained through Geneticin selection. At 50 days posttransfection, episomal DNA was isolated through Hirt extraction and the copy number was established through qPCR with LCR primers. (D) RNA was extracted from the cells in B and amplified with primers to E1̂E4. (E) Total DNA was isolated from HaCaT cells at 1 and 2 days postinfection with quasivirus, amplified with primers to LCR, and normalized to the WT genome. (F) Hirt extraction of HaCaT cells infected with quasivirus and stably maintaining genome for 50 days. (G) Same as E except NIKS cells were infected for 14 days. (H) Episomal DNA from 14-day NIKS infection isolated through the exonuclease V method. (I) Controls for exonuclease digestion of part H. Linear genomic actin DNA should be completely digested by exonuclease, while circular mitochondrial DNA is more resistant to digestion. Each graph is representative of multiple infections, and means are expressed ± SEM.
