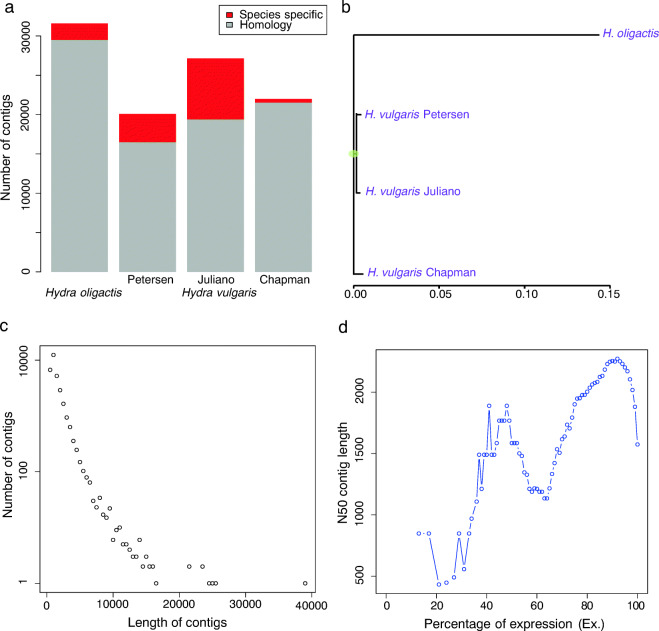
Fig. 1.
De novo transcriptome assembly of H. oligactis. a The number of contigs in H. oligactis and H. vulgaris transcriptomes. About 93.4% of assembled H. oligactis contigs shows homology to H. vulgaris. Only 2% of the mRNAs published in NCBI by Chapman et al. was missing, which reflect putative species-specific genes. b Phylogenetic tree of Hydra transcriptomes. The H. vulgaris from Chapman et al. originated from a different subgroup of H. vulgaris. c Length distribution of final assembled contigs. 60.3% of contigs were shorter than 1 kb, while only 0.2% were longer than 10 kb. d The distribution of N50 of the contigs with an expression value that represented percentage of the total expression data (ExN50). About 90% of the expression data is represented by contigs of an N50 of 2255 bp