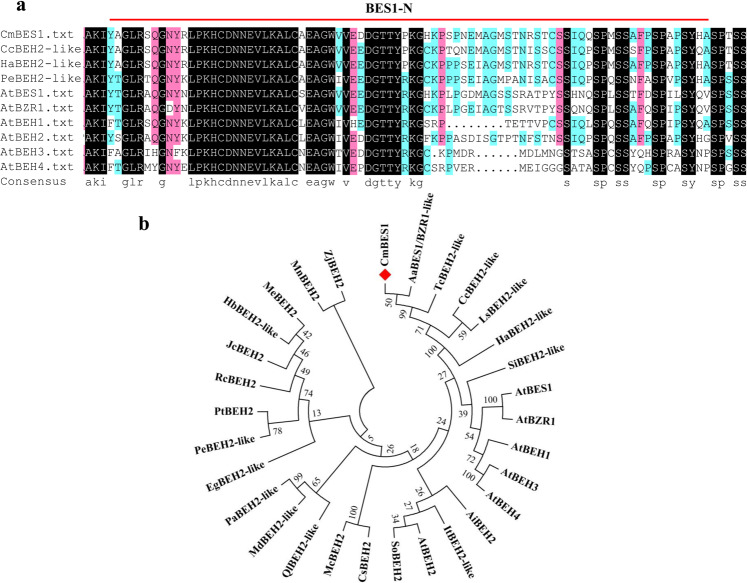
Fig. 1. Deduced amino acid sequence comparison and phylogenetic analysis of CmBES1.
a Amino acid sequence alignment of CmBES1 with BES1 sequences from various plant species. The single red lines indicate the conserved BES1-N region in BES1-like proteins. The black color indicates 100% identity; red, 75% identity; and blue, 50% identity. The sequences compared to CmBES1 were Arabidopsis thaliana AtBES1 (AT1G19350), AtBZR1 (AT1G75080), AtBEH1 (AT3G50750.1), AtBEH2 (AT4G36780), AtBEH3 (AT4G18890), and AtBEH4 (AT1G78700); Helianthus annuus BES1/BZR1 homolog protein 2-like (HaBEH2-like, LOC110908961); Cynara cardunculus BES1/BZR1 homolog protein 2-like (CcBEH2-like, LOC112512677); and Populus euphratica BES1/BZR1 homolog protein 2-like (PeBEH2-like, LOC105107647). b Phylogenetic tree comprising the following CmBES1 and BES1 family proteins: Arabidopsis thaliana AtBES1 (AT1G19350), AtBZR1 (AT1G75080), AtBEH1 (AT3G50750.1), AtBEH2 (AT4G36780), AtBEH3 (AT4G18890), and AtBEH4 (AT1G78700); Arachis ipaensis AiBEH2 (XP_016201823.1); Artemisia annua AaBES1/BZR1-like (PWA37330.1); Cucumis sativus CsBEH2 (XP_004143497.1); Cynara cardunculus CcBEH2-like (LOC112512677); Eucalyptus grandis EgBEH2-like (NP_001306904.1); Helianthus annuus HaBEH2-like (LOC110908961); Hevea brasiliensis HbBEH2-like (XP_021646974.1); Ipomoea triloba ItBEH2-like (XP_031095547.1); Jatropha curcas JcBEH2 (XP_012079081.1); Lactuca sativa LsBEH2-like (XP_023759843.1); Malus domestica MdBEH2-like (XP_008338783.2); Manihot esculenta MeBEH2 (XP_021632984.1); Momordica charantia McBEH2 (XP_022132946.1); Morus notabilis MnBEH2 (XP_024020611.1); Populus euphratica PeBEH2-like (LOC105107647); Populus trichocarpa PtBEH2 (XP_002310201.1); Prunus avium PaBEH2-like (XP_021809451.1); Quercus lobata QlBEH2-like (XP_030949816.1); Ricinus communis RcBEH2 (XP_002525100.1); Sesamum indicum SiBEH2-like (XP_011097209.1); Spinacia oleracea SoBEH2 (XP_021835401.1); Tanacetum cinerariifolium TcBEH2-like (GEY43666.1); and Ziziphus jujuba ZjBEH2 (XP_015890203.1). The phylogenetic tree was constructed using the neighbor-joining method and bootstrap test with 1000 replicates. The divergence of each branch is indicated by the bootstrap values