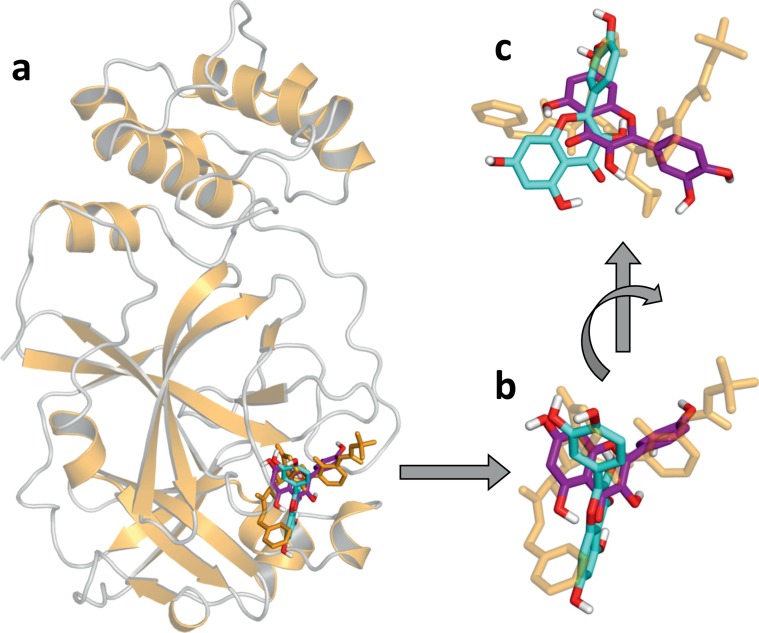
Fig. 9.
Molecular docking of quercetin to 3CLpro. Structure of (a) unliganded 3CLpro (from entry 6Y2E of the PDB; in ribbon representation, yellow) and α-ketoamide inhibitor bound in the active site (from entry 6Y2F; in stick representation, yellow) [22]. The liganded protein was superimposed by a least square fit on the Cα atoms of the liganded one, and only the latter is shown. (b) and (c) Detail of the 3CLpro active site, with the two best docking poses obtained using as receptor the structure 6Y2E (cyan) and 6Y2F (purple). Polar hydrogen atoms are explicitly shown (white) and oxygen atoms are also coloured (red).