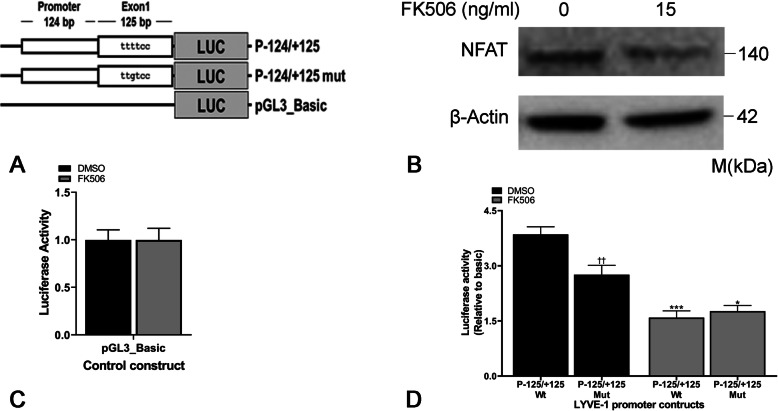
Fig. 4.
Luciferase activity of LYVE-1 promoter expression constructs. Schema of pGL3_Basic vector, and wildtype and mutant LYVE-1 luciferase promoter constructs transfected into HEK-293 T cells (a). Endogenous NFAT expression in HEK-293 T cells (b). Luciferase activity of vehicle (DMSO) and FK506 treated HEK-293 T cells transfected with (c) pGL3_Basic vector and (d) wildtype and mutant LYVE-1 promoter constructs. Each luciferase data is normalized to internal Renilla control. Data for LYVE-1 promoter constructs are presented relative to the pGL3_Basic vector luciferase activity. Average (± standard deviations as error bars) of four independent experiments are presented; each independent experiment for western blotting consisted of three technical replicates. *** p < 0.0001 (between P-125/+ 125 wildtype DMSO vs. FK506), * p < 0.05 (between P-125/+ 125 mutant DMSO vs. FK506), †† p < 0.001 (between P-125/+ 125 wildtype DMSO vs. P-125/+ 125 mutant DMSO) by 2-way ANOVA (Tukey’s multiple comparison) test