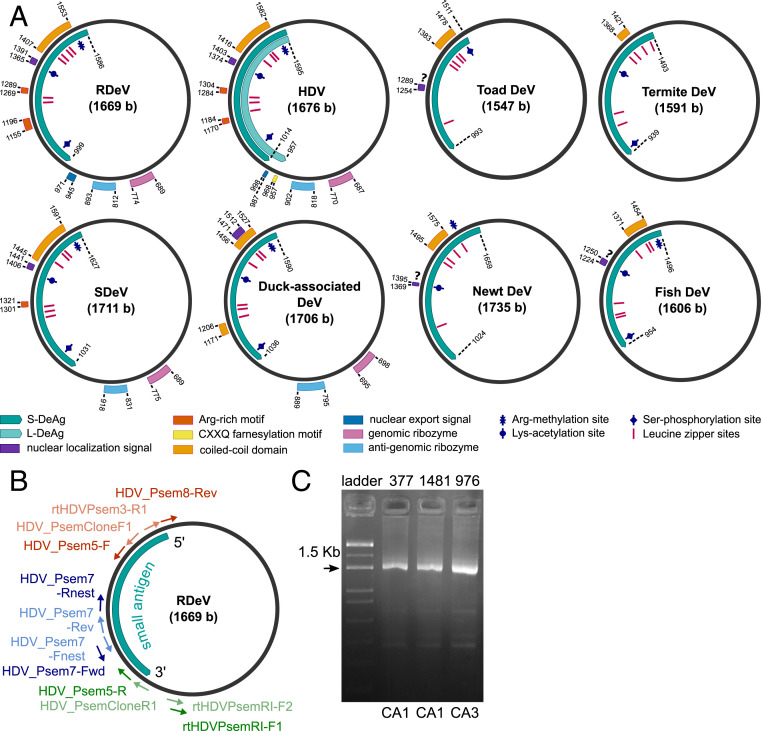
Fig. 1.
(A) Genome organization of human HDV, RDeV, SDeV, duck-associated DeV, newt DeV, toad DeV, fish DeV, and termite DeV. Delta antigen-coding regions, functional and structural motifs, as well as post-translational modification sites. (B) Primer-binding positions for the circularization assays shown in their locations on the RDeV genome. Primer names and sequences are listed in SI Appendix, Table S2. Blue, circularization assay 1 (CA1); red, circularization assay 2 (CA2); green, circularization assay 3 (CA3). External primers are shown in dark color, and nested primers in light color. The S-RDeAg ORF is shown for orientation. (C) Exemplary results from CA1 and CA3 obtained from total RNA blood extracts of three RDeV RNA-positive P. semispinosus. Fragment sizes are 1,608 bp for CA1 and 1,532 bp for CA3; label correspondence to RDeV GenBank accession numbers: 377, MK598006; 1481, MK598003; 976, MK598009.