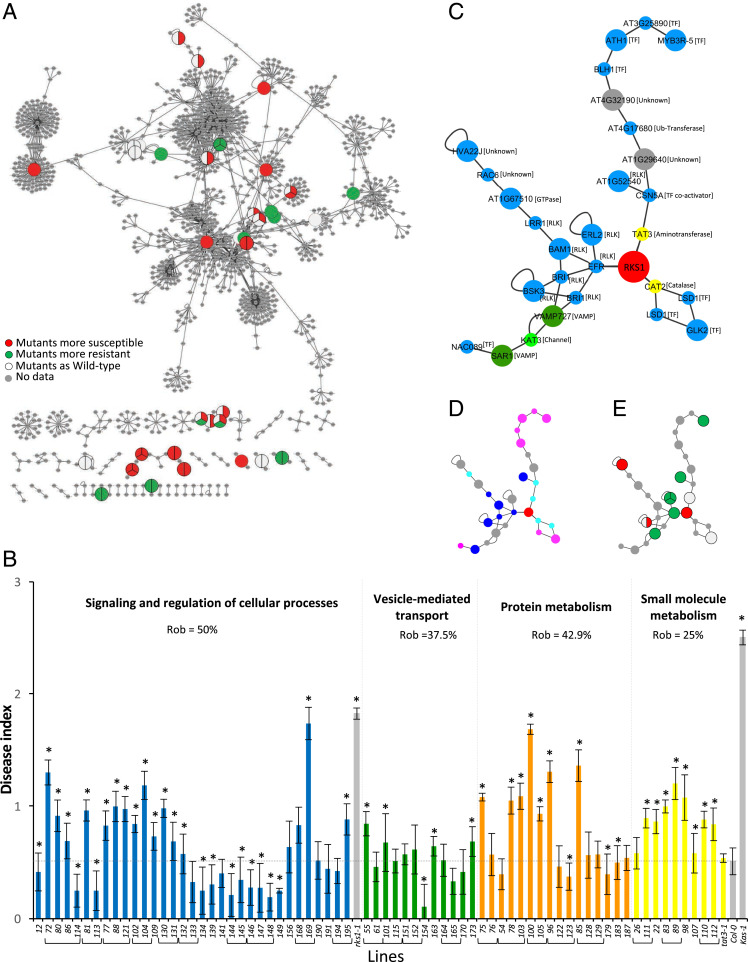
Fig. 5.
Evaluation of robustness of the RKS1-dependent network by phenotypic characterization of mutants corresponding to some network components. (A) The RKS1 protein–protein interaction network plotted with Cytoscape showing the components for which mutants have been phenotyped in response to Xcc. Each circle represents the mutant phenotype corresponding to the protein component of the network: red indicates mutants significantly more susceptible than the wild type accession, green more resistant, and white not affected. No mutant was tested for genes represented in gray. (B) Disease index at 7 dpi after inoculation with a bacterial suspension adjusted to 2.108 cfu/mL of mutants corresponding to genes belonging to the different functional groups. Mutants belonging to “Signaling and regulation of cellular process” (blue), “Vesicle-mediated transport” (green), “Protein metabolism (ubiquitination/proteasome)” (orange), and “Small molecule metabolism” (yellow) are identified by numbers (SI Appendix, Table S3 shows correspondence to gene accession numbers). Mutants corresponding to the same gene have been grouped. Means were calculated from 4 to 24 plants (*Kinetic modeling deference with Col-0 time course, 0 = P value > 0.05 and 1 = P value ≤ 0.05). (C–E) A signaling subnetwork was extracted from the RKS1 protein–protein interaction network. (C) Subnetwork with protein functional groups (blue, signaling and regulation; yellow, small molecule metabolism; green, small molecule transport; dark green, vesicle-mediated transport; grey, others) and the protein molecular functions. Large circles indicate proteins encoded by genes from the 268 DEGs, and little circles indicate proteins from Y2H or BioGRID candidates. (D) Subnetwork with the protein subcellular localization (dark blue, plasma membrane; pale blue, cytoplasm; pink, nucleus; grey, others). (E) Subnetwork with the insertional mutant phenotypes (red, mutants more susceptible than the wild-type; green, mutants more resistant than the wild-type; white, mutants as the wild-type; grey, not determined).