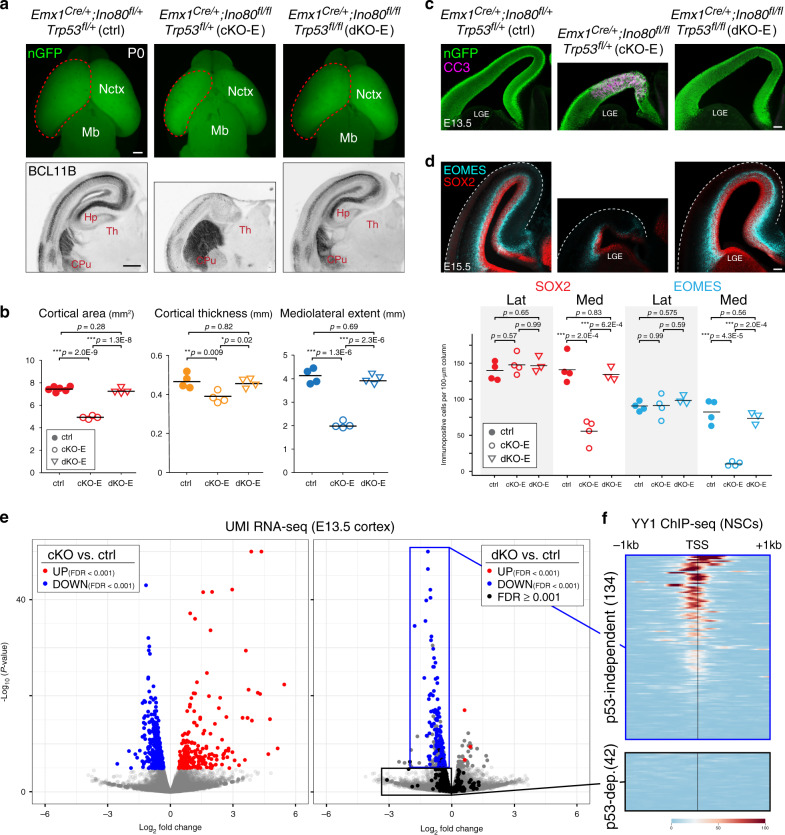
Fig. 5. Trp53 co-deletion rescued Ino80 phenotypes and revealed mechanistically distinct Ino80 roles.
a Dorsal view of P0 whole-mount brains and BCL11B immunostaining (black) of P0 coronal sections. Co-deletion of Trp53 with Ino80 (dKO-E) rescued major Ino80 deletion (cKO-E) phenotypes, including microcephaly, severe hippocampal hypoplasia, and disrupted neocortical lamination. Sample measurements quantified in b are indicated (ctrl cortical area: n = 6, all other measurements: n = 4 animals). b Cortical area (red), thickness (yellow), and mediolateral extent (blue) were restored in dKO-E and not significantly different compared with ctrl (data are mean, one-way ANOVA with Tukey’s post hoc test, ctrl cortical area: n = 6, all other measurements: n = 4 animals). c CC3 immunostaining (magenta) revealed no increase in apoptosis in dKO-E E13.5 neocortex (n = 3 animals). d The number of SOX2 + (red) and EOMES + (cyan) NPCs in E15.5 dKO-E cortex were restored to levels not significantly different compared with ctrl (data are mean, one-way ANOVA with Tukey’s post hoc test, ctrl: n = 4, cKO-E: n = 4, dKO-E: n = 3 animals). e UMI RNA-seq volcano plots comparing cKO-E with ctrl, and dKO-E (n = 7 animals) with ctrl E13.5 cortex. In the cKO-E versus ctrl comparison (left panel), differentially regulated genes are indicated (upregulated, red dots; downregulated, blue dots). The same genes are labeled in the dKO-E versus ctrl comparison (right panel). Genes that remained differentially regulated (FDR < 0.001) following Trp53 co-deletion are indicated by dots of the same color. Genes that lost differential expression (FDR ≥ 0.001) are indicated by black dots. The vast majority of cKO-E-upregulated genes (202/205) were rescued by Trp53 co-deletion, indicating that their upregulation was p53-dependent. About one-third of cKO-E-downregulated genes (134/418), however, remained significantly downregulated in dKO-E, consistent with p53-independent gene regulation by Ino80. f Intersectional analysis of the 134 p53-independent downregulated genes with published ChIP-seq data from mouse neural stem cells (NSCs) revealed an enrichment of genes bound at their transcriptional start site (TSS) by transcription factor YY1, a known binding partner of INO80. This enrichment was absent from the 42 cKO-E downregulated genes that were reversed by Trp53 co-deletion. Scale bar: 500 μm in a; 100 μm in c, d.