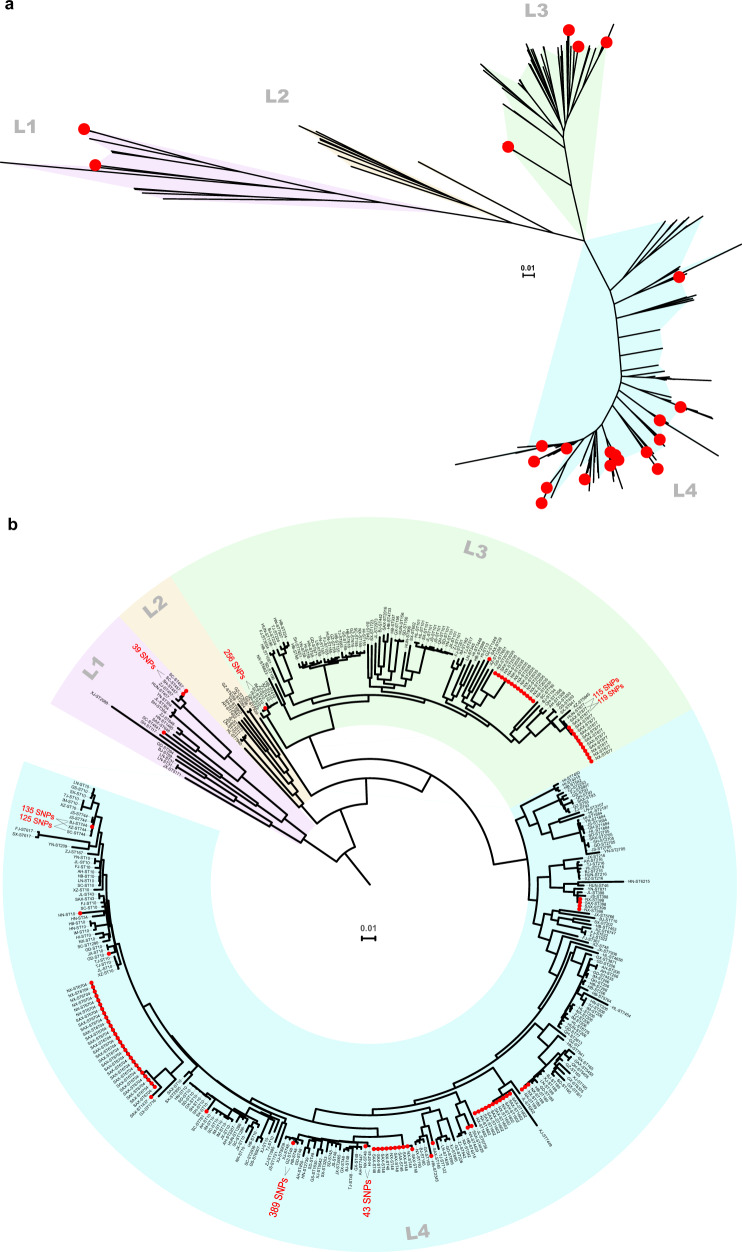
Fig. 6. High genetic propensity of tet(X4)-harbouring E. coli in spreading into humans.
Phylogenetic trees of the 95 tet(X4)-positive E. coli isolates together with 287 publicly available draft genomes of tet(X4)-negative E. coli (PRJNA400107) from healthy humans across China (Neighbour-Joining trees constructed based on 166,544 core-genome SNPs). Red dots represent the 95 isolates positive for tet(X4). a An unrooted tree showing the four Bayesian lineages. b A circular tree showing the corresponding information and SNP differences between isolates that are positive or negative for tet(X4). Abbreviations: AH, Anhui; BJ, Beijing; FJ, Fujian; GS, Gansu; GD, Guangdong; GX, Guangxi; GZ, Guizhou; HI, Hainan; HB, Hubei; HUN, Hunan; HN, Henan; HL, Heilongjiang; JL, Jilin; JS, Jiangsu; JX, Jiangxi; LN, Liaoning; IM, Inner Mongolia; NX, Ningxia; QH, Qinghai; SD, Shandong; SX, Shanxi; SAX, Shaanxi; SH, Shanghai; SC, Sichuan; TJ, Tianjin; XZ, Xizang; XJ, Xinjiang; Yunnan; ZJ, Zhejiang.